Shutterstock.com
Sequencing the first human genome took more than a decade and cost just over £1.5bn. Fast-forward 14 years and a whole genome can now be sequenced in a matter of days for a few hundred pounds.
“We’ve come a long way,” says Joanne Hackett, commercial director of Genomics England. “Today, thanks to next-generation sequencing, we can run hundreds of samples in parallel and read all 3.2 billion letters of DNA in less than 3 days at [a cost of] around £700.”
The Human Genome Project (HGP), which ran from 1990 to 2003, was a publicly funded project to sequence the first human genome. It was supported by the Medical Research Council and the Wellcome Trust in the UK, and the Department of Energy and US National Institutes of Health in the United States. Since the HGP ended, further projects have built on its foundations to reveal, bit by bit, how our genomes influence our growth, development and health[1]
. In September 2003, the National Human Genome Research Institute (NHGRI) in the United States launched a project named ENCODE (the Encyclopedia Of DNA Elements), which found that a biochemical function could be assigned to 80% of the human genome, a striking discovery since it was initially thought that a vast proportion of the genome was simply “junk”.
Next came the 1,000 Genomes Project, an international research effort launched in 2008, which was the first project to sequence the genomes of a large number of people from different ethnic groups to construct a comprehensive resource on human genetic variation. Following on from this, in 2010, the Wellcome Trust launched the UK10K study, which looked at 10,000 human genomes from individuals living in the UK — 4,000 healthy people and 6,000 people currently living with a disease of suspected genetic causes, such as severe obesity or congenital heart disease.
Then, in late 2012, then prime minister David Cameron launched a challenge to sequence 100,000 genomes by 2017, a deadline that was extended until the end of 2018[2]
. To help things along, the Department of Health set up Genomics England to run the 100,000 Genomes Project. As of 7 August 2017, 32,642 genomes had been sequenced.
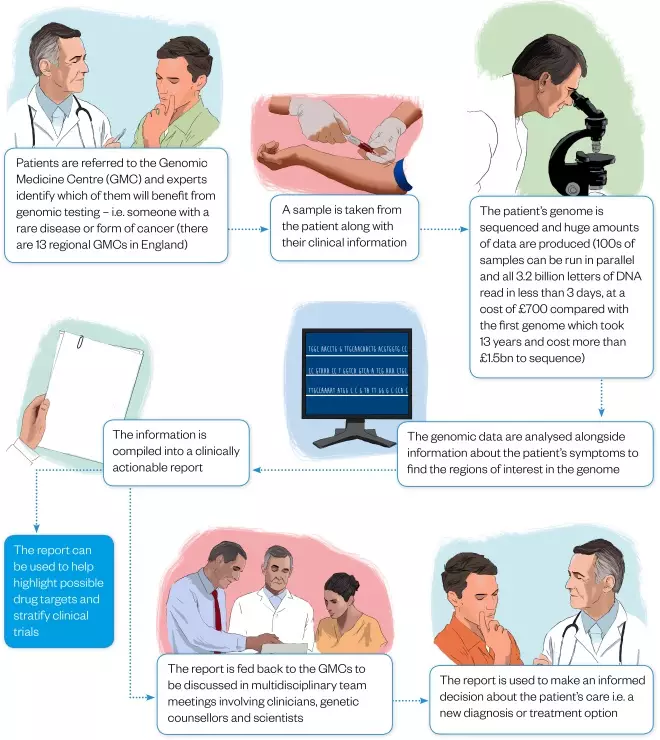
Figure 1: From diagnostic odyssey to diagnosis
Illustrations: Javier Maria Trigo Gonzalez
As of August 2017, more than 32,000 genomes have been sequenced, making the UK’s 100,000 Genomes Project one of the largest national sequencing projects of its kind in the world.
Each person’s genome is made up of a unique sequence of DNA that contains the instructions for their growth and development. Differences in our DNA determine our unique characteristics but can also underlie certain diseases. Sequencing the genomes of many individuals enables researchers to gain a better understanding of these diseases to develop treatments and make new diagnoses. Variations in a person’s genome can also affect their individual response to a drug, and genome sequencing can help match patients to the most appropriate drug, work out the best dose and predict whether they are likely to suffer adverse effects — a field called pharmacogenomics.
The 100,000 Genomes Project is driving the move away from a ‘one size fits all’ approach to treatment, towards personalised, or precision, medicine, whereby each patient is treated according to their individual genome sequence. Genetic information is also being used by pharmaceutical companies to help inform drug discovery and development, and reduce the associated time and cost (see Panel 1). As genomics becomes a reality for the NHS, pharmacists and their healthcare colleagues will need to prepare to face new challenges and transformations in the way care is thought about and delivered.
Genome sequencing
The 100,000 Genomes Project is one of the largest national sequencing projects of its kind in the world and is focused primarily on two therapeutic areas: cancer, which involves sequencing the DNA from both the patient’s tumour and their healthy cells to compare the two; and rare diseases, which involves sequencing the genomes of the affected person and the genomes of two close blood relatives.
“With the consent of our participants, we are able to link genomic data with additional health data from GP, hospital and other sources such as national disease registries,” explains Hackett. “We use this information all together to enable better clinical interpretation of the genomes.”
With the consent of our participants, we are able to link genomic data with additional health data from GP, hospital and other sources such as national disease registries
Patients with rare diseases may spend a long time in a ‘diagnostic odyssey’— a cycle of tests, hospital visits and unanswered questions that, for some patients, can last years. A diagnosis often brings these patients relief and hope by enabling the next steps in their care. “You can put the family’s mind at rest, help with practical matters of family planning and in some cases there are treatments available,” explains David Bentley, vice-president and chief scientist at Illumina, Genomic England’s sequencing partner for the 100,000 Genomes Project.

Source: Courtesy, Illumina
David Bentley, vice-president and chief scientist at Illumina, Genomic England’s sequencing partner for the 100,000 Genomes Project, says that a diagnosis often brings relief for patients with rare diseases and hope by enabling the next steps in their care
“A rare disease that hits very early can be a fairly straightforward metabolic defect that’s easy to interpret, and Genomics England has already given examples of how changing diet or providing certain treatments or supplements can make a difference. There’s real actionable information coming out,” he says.
Illumina’s sequencing work is being carried out in a purpose-built centre on the Wellcome Genome Campus, home of the Sanger Institute, the European Bioinformatics Institute (EBI) and a number of new biotechnology companies, in Hinxton, near Cambridge, UK. Bentley has been involved in the project from the start.
“No one had ever done anything on that scale so it wasn’t simple but it was smooth,” says Bentley, explaining how Illumina created a clear path from sequencing the DNA to delivering the annotation, which involves marking where the genes in the DNA sequence start and stop, and also where other interesting regions are in the sequence.
“It took a great deal of work,” he says. “One of the challenges was to enable the technology to go on developing, without disturbing the consistency of the data.
“We worked closely with Genomics England on that.”
In 2016, it was announced that Illumina would also be working with Genomics England to develop a set of tools for researchers and clinicians in regional NHS genomic medicine centres (GMCs) to access genomic information and reports more easily, helping to bridge the gap between science and healthcare. Genomics England also has contracts with Congenica, Fabric Genomics, Nanthealth and WuxiNextcode, which are all providing complementary clinical interpretation services in the project.

Source: Wellcome Trust Sanger Institute / Genome Research Limited
Illumina’s sequencing work is being carried out in a purpose-built centre on the Wellcome Genome Campus, home of the Sanger Institute, the European Bioinformatics Institute (EBI) and a number of new biotechnology companies, in Hinxton, near Cambridge, UK
Data interpretation
Once the genome sequencing is completed by Illumina, work then begins to analyse the data and find out what it all means.
“The sequencing part [of the project] is relatively easy and fast,” explains Hackett. “The challenge lies in data analysis and interpretation, which take longer.”
The sequencing part [of the project] is relatively easy and fast. The challenge lies in data analysis and interpretation, which take longer
Hackett explains that we still don’t yet know what all of the regions of the human genome mean and what they are responsible for.
“There’s currently no gold standard pipeline or platform to be able to analyse the human genome, and definitively mark up the one or few changes in DNA that might be responsible for a rare condition or cancer,” she says. “Much of the interpretation requires skilled bioinformaticians, manually analysing genomic data, along with information about the patient’s condition, to pick out the needle in a haystack,” she adds.

Courtesy of Genomics England
Joanne Hackett, commercial director of Genomics England, says that we still don’t yet know what all the regions of the human genome mean and what they are responsible for
Genomics England has its own research arm known as the Genomics England clinical interpretation partnership (GeCIP), which brings together more than 2,500 clinicians, trainees and researchers who are organised into specialities, such as breast cancer or cardiovascular disease.
The role of the GeCIP is to improve understanding of genomic medicine and its application to healthcare, improve understanding of disease and lead the way towards developing new diagnostics and treatments.
However, Genomics England has also taken on external interpretation providers to help deal with the vast amounts of sequencing data. The first to be chosen for the job was Congenica, a spin-off from the Sanger Institute with the mission of revolutionising medical diagnosis and integrating genomics into mainstream healthcare.
To do this, Congenica has developed a genome analytics and interpretation software platform called Sapientia. The cloud-based platform enables its users, who range from medical practitioners to genetic counsellors across the world, to take a patient’s genomic data, combine it with their clinical symptoms, and then, through various algorithms and prediction tools built within Sapientia, rank that individual’s variants as either ‘pathogenic’, ‘non-pathogenic’ or ‘likely to be pathogenic’.
According to Shikha O’Brien, Congenica’s chief business officer, this process takes mere minutes. “A team of clinical scientists at Congenica [then] reviews and refines the report,” she says.
Once the reports are ready, they are then fed back to Genomics England before being passed on to the most appropriate NHS GMC to be used to help make informed decisions about a patient’s care.
“We’re not going to look for the easy diagnosis and keep people in that horrible diagnostic odyssey,” explains Alan Martin, head of innovation and data science at Congenica, “if you’re phoning up NHS 111 you’re getting a flowchart that’s been built up from evidence.

Source: Courtesy, Alan Martin
Alan Martin, head of innovation and data science at Congenica, says the company is working with drug companies to help them better stratify their trials
“What we’re doing in this process is creating your flowchart — not the one for everyone, but the one for you. This is what Genomics England and Congenica are all about.”
What we’re doing in this process is creating your flowchart — not the one for everyone, but the one for you
Into the healthcare system
There are 13 regional GMCs across England; Scotland and Northern Ireland have set up their own separate but affiliated programmes and there are plans for one in Wales too.
One role of the GMCs is to identify those patients, many of whom are referred from secondary care, that might benefit most from genomic testing — individuals with rare diseases or different forms of cancer — and then provide them with information about what the testing involves. Once an individual’s genome has been sequenced and analysed, the results are compiled into a clinically actionable report, which is fed back to the GMCs for them to discuss in multidisciplinary meetings before informing patients.
The GMCs are also closely involved in the education and training of healthcare professionals. In Manchester, the GMC runs genome café events, where anyone from patients to clinicians can drop in to talk to experts about genomics.
“One of the challenges in genomics is making sure that all those doctors, nurses, pharmacists and medical students know what genomics is,” explains William Newman, clinical lead at the Greater Manchester GMC. “It’s a much larger undertaking than I think people considered initially.”
One of the challenges in genomics is making sure that all those doctors, nurses, pharmacists and medical students know what genomics is. It’s a much larger undertaking than I think people considered initially
To ensure that NHS staff across the country have the knowledge, skills and experience required, the Genomics Education Programme (GEP), a £20m, three-year programme, was set up by Health Education England (HEE).
“Formal and informal education materials and training courses are available through the GEP,” explains Ellen Brennan-Rist, a pharmacist and genomics champion for the West of England GMC. “Pharmacists may wish to find out what is happening in their area and how they can get involved by visiting the Genomics England website or that of their local GMC.”

Courtesy of Ellen Brennan-Rist
Ellen Brennan-Rist, a pharmacist and genomics champion for the West of England genomic medicine centre (GMC), says that pharmacists can find out what is happening in their area and how they can get involved by visiting the Genomics England website or the website of their local GMC
At the highest level, HEE also funds a genomic medicine master’s programme being delivered at 10 universities across England with funding available for 550 places for NHS staff over the 3 years.
Emma Groves, who registered as a hospital pharmacist in 2015, was inspired to apply for the master’s at Newcastle University after attending a lecture that promoted the integration of genomic technologies within the NHS.
“Genomics has the potential to improve patient outcomes through appropriate stratification of medicines and pharmacists should be part of this exciting development,” says Groves, whose fellow students on the course include doctors, biomedical students, GPs and a clinical scientist, as well as pharmacists.
Genomics has the potential to improve patient outcomes through appropriate stratification of medicines and pharmacists should be part of this exciting development
Newman, a doctor with a background in clinical genetics, predicts that in the future there will be a drive to mainstream sequencing technology so that clinicians in all other areas can have access to the testing. “But they need to know what they’re asking for, how to interpret the results and how to apply it in a clinical setting, so I think the GMCs will become a place for people to seek expert advice or opinion,” he says.
The GMC in Greater Manchester, which covers a population of around 3.5 million, is already expanding its services across hospitals in and around the region to ensure that patients don’t have to travel, as well as raise awareness about what genetics has to offer.
Newman highlights that the 100,000 Genomes Project will lead to greater equity of access to genetic testing. “I think we’ll end up finding out how best to use it in the clinical pathway for an individual patient, and I think it’ll be used in such a way that individuals, wherever they live in England, will have much better access to that testing.”
Becoming mainstream
On 3 July 2017, Dame Sally Davies, chief medical officer, published her annual report for 2016, entitled ‘Generation genome’, in which she says that genomics is already with us — the issue is that not everyone can access it[3]
. Her “genomic dream”, as outlined in the annual report, is for whole-genome sequencing to be widely adopted across the NHS, where appropriate, to speed up diagnosis and treatment for all patients.
As part of this shift, pharmacists need to prepare. From Newman’s position as a researcher and clinician, he believes that pharmacists, as experts in medicines, will have an important role to play. “Pharmacists will be using [genomic] information when they see a patient on the ward, or passing through, who has been prescribed a certain medication, to find out if the patient has had the right genetic test; and has the pharmacist got the right genetic information, which means that a medication is appropriate for the patient?”

Courtesy of William Newman
William Newman, clinical lead at the Greater Manchester Genomic Medicine Centre, believes the 100,000 Genomes Project will lead to greater equity of access to genetic testing
Brennan-Rist says that pharmacists need to embrace the move towards a more holistic and individual view of disease management. “Personalised medicine is already happening, for example, in cancer therapy, where genetic changes can guide which treatments are more likely to be effective and less likely to cause adverse effects,” she says.
“In a couple of years, whole-genome sequencing will become mainstream clinical practice… it will provide even more patient-specific data, on which diagnosis and treatment choices are made,” she adds.
In a couple of years, whole genome sequencing will become mainstream clinical practice… it will provide even more patient-specific data, on which diagnosis and treatment choices are made
Groves believes that pharmacists will need to keep up to date with the genomic basis of drug efficacy, adverse drug reactions and how this information can impact prescribing.
“As pharmacists, there will [also] be opportunities to be more involved with the genomic-related research and support patients through genetic counselling and pharmacogenomics-related advice,” she adds.
With just over a year left before the 100,000 Genomes Project deadline, healthcare is already being transformed by genomics. And with sequencing set to become cheaper and faster, this transformation is on an upward trajectory. “There is a really interesting megatrend here, which is moving from individual knowledge, experience and skills towards evidence-based healthcare,” says Martin. “It is a fundamental change for the NHS and a cultural shift.”
Panel 1: Drug discovery and development
Genomic information is used to liaise with pharmaceutical companies to find out what they are currently working on, highlight possible drug targets and help them get medicines to market quicker by influencing clinical trial design. “[Congenica is] working with drug companies to help them better stratify their trials,” explains Alan Martin, the company’s head of innovation and data science. “Having a database of biological knowledge is where we need to aim ourselves.”
Using genomics to drive the selection of better drug targets is the primary aim of Open Targets, a pre-competitive consortium composed of four collaborative partners, GlaxoSmithKline (GSK), Biogen, the Sanger institute and the European Bioinformatics Institute.
“We have published data to show that if you have good genetic information you are almost twice as likely to have a successful launch of a [drug] later on,” confirms Philippe Sanseau, head of computational biology at GSK.
“When we fail, we tend to fail at late stage, which is very expensive. Using genetics reduces attrition, saves money, [identifies] better targets and is maybe faster as well.”
As its name suggests, all information generated by Open Targets, which focuses primarily on the areas of oncology, neurodegeneration and immunology, is shared across the four partners and is openly accessible to all. This includes the public, who can use the Open Targets online platform to answer questions such as ‘Which genes are associated with disease?’ Together, the four partners come up with areas of investigation, which they discuss, tweak and modify together until a molecule is created. After that, the project moves into the competitive arena.
“[It is] true collaborative, open working. [It] takes a lot longer and a lot more money to do this alone. We hope we can benefit ourselves and other researchers [through this approach],” explains Sanseau.
Longer term, he says, one of the biggest impacts is on new drug development. “We get a better understanding of the disease population we want to tackle and with the confidence that your drug will be efficacious and safe for the right disease and the right population, you can save money for healthcare systems across the world.”
References
[1] yourgenome.org. How is the completed human genome sequence being used? Available at: http://www.yourgenome.org/stories/how-is-the-completed-human-genome-sequence-being-used (accessed August 2017)
[2] Genomics England. The 100,000 Genomes Project. Available at: https://www.genomicsengland.co.uk/the-100000-genomes-project/ (accessed August 2017)
[3] Davies S. Annual report of the chief medical officer 2016. Generation Genome. Department of Health; 2017. Available at: https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/624628/CMO_annual_report_generation_genome.pdf (accessed August 2017)