Shutterstock.com
Everyone strives to define themselves as an individual, distinguished from the crowd and defined by their own desires and experiences. We are exactly this: biologically unique, and (with the exception of identical twins) the owners of a distinct pattern of DNA unlike anyone else’s (Box 1: ‘Creating an individual’). It therefore seems antiquated, in a world more accepting of difference than ever before, to accept a universal, generalised idea of how a person will respond to a drug.
Pharmacogenomics involves the investigation of the effects of genetic variation, the differences in our genome, on drug response. Pharmacogenomics aims to improve the safety and effectiveness of drug treatment by using genetic information to inform prescription decision making, similarly to how renal function is taken into account when prescribing drugs that are eliminated by the kidneys. In this process, genetic variants that influence drug response are identified, characterised and then used to stratify patients into subgroups that receive subgroup-specific, genotype-informed prescriptions. Therefore, pharmacogenomics is a cornerstone in the ongoing endeavour for precision medicine, also known as stratified or personalised medicine.
The term ‘pharmacogenetics’ was first coined by the German geneticist Friedrich Vogel in 1959[1]
and relates to the study of inherited genetic differences in drug metabolic pathways that can affect individual responses to drugs, both in terms of therapeutic effect as well as adverse effects. However, the concept that innate factors can affect drug response dates back as far as Pythagoras[2]
. Pharmacogenomics is often used interchangeably with pharmacogenetics, and was first used in 1997[3]
.
Current prescribing approaches include a degree of individual tailoring based on factors such as age, weight, biochemical markers and co-morbidities. However, despite these considerations some patients do not respond in the expected manner. The number of patients who respond beneficially to a given drug can be anything between 25% and 80%[4]
. Furthermore, around 6% of hospital admissions are related to an adverse drug reaction (ADR)[5]
, and drug tolerability issues are associated with poor medication adherence[6]
. Pharmacogenomics aims to offer clinical decision support to improve the benefit–risk profile of prescribed medications.
Box 1: Creating an individual
The human genome is the instruction manual for building a person. DNA, the language of the text, comprises sequences of nucleotide bases that are split into sections known as genes. Each gene is responsible for encoding the information to create a specific protein.
The genome remains largely similar from person to person with small variations, known as polymorphisms, present in everyone. These polymorphisms are either germ-line (inherited) or somatic (acquired).
An allele is a variant form of a gene caused by a polymorphism. If a specific allele occurs in less than 1% of a population, it is typically termed a mutation. While many polymorphisms do not significantly alter gene function, some affect gene expression or the structure and ultimately the activity of the encoded protein. These are of particular interest with regard to diagnostics and therapeutics.
These instructions are also influenced by ‘epigenetic’ modifications that can influence gene expression via mechanisms such as switching off an activated gene. These modifications can be due to factors such as age, lifestyle or disease and can result in potential alterations to the expression of phenotypes without changing the underlying genotype[7]
.
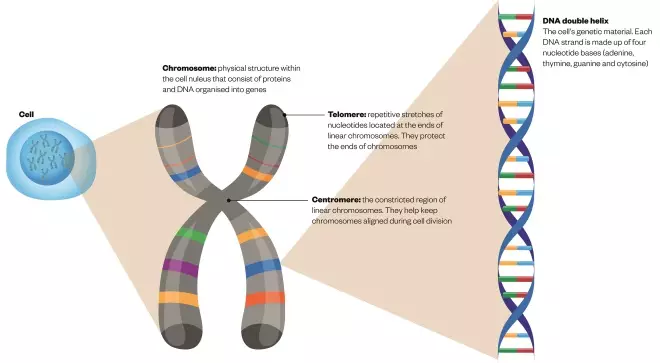
What makes up a chromosome?
Source: The Pharmaceutical Journal
DNA, or deoxyribonucleic acid, is the hereditary material in humans and almost all other organisms. It comprises sequences of nucleotide bases that are split into sections known as genes
The genetics of drug response
It is estimated that more than 97% of people carry at least one variant in a gene that can influence drug response by disturbing either the pharmacokinetics or the pharmacodynamics of a drug[8]
. For example, some genetic variants impact on drug pharmacokinetics by affecting genes involved in drug ADME: absorption, distribution, metabolism and excretion, including drug-metabolising enzymes (e.g. cytochrome P450 isoforms, CYPs) and drug transporters. Genetic variants can also influence both a drug’s on-target and off-target pharmacodynamic effects.
The majority of ADRs that lead to hospital admission are type A reactions (over 80%). These are predictable from knowledge of a drug’s pharmacology and usually result from excessive drug activity through increased systemic exposure and/or increased sensitivity of the target site to the drug[9]
.
The identification of mutations is also being used to design new drugs that must be used in patients with that mutation. This is most relevant in cancer medicine, where pharmacogenomics currently has, and will have, increasing relevance. For example, vemurafenib was designed to inhibit the V600E mutation in the BRAF gene in metastatic malignant melanoma[10]
. The use of targeted therapies, such as trastuzumab, in HER2-positive breast cancer is another example[11]
. Similar approaches are now also being used for the germline genome. Ivacaftor is a novel therapeutic licensed for patients with cystic fibrosis (CF) carrying certain pathogenic mutations in the CF faulty protein, CFTR. Ivacaftor binds to and partially restores the activity of CFTR, but only works in patients carrying specific CFTR mutations, and thus requires a knowledge of the genotype before prescribing[12]
.
P450 genes
The effect of environmental influences on CYP metabolising activity and drug response is extensively covered in the pharmacy curriculum (e.g. drug–drug and drug–food interactions). However, a large number of CYP genes are polymorphic and, therefore, the influence of genetics on variation in response should not be underestimated.
The term ‘metaboliser phenotype’ is often used when predicting the activity of a drug-metabolising enzyme in an individual patient, based on the gene alleles the patient carries and prior knowledge of the typical activity level of each allele. However, some enzymes, such as CYP2D6, lead to four different activity phenotypes: poor metaboliser, intermediate metaboliser, extensive (normal) metaboliser and ultra-rapid metaboliser[13]
.
More than 100 different CYP2D6 allelic variants have been recorded to date[14]
. Around 6–10% of Caucasians are considered poor metabolisers; however, the incidence varies among ethnic groups with less than 1% of Asians and 2–5% of African–Americans lacking the enzyme[15]
. CYP2D6 is responsible for the metabolism and/or activation of around 25% of prescribed drugs[16]
, including for example, codeine. Codeine is a prodrug and a small proportion (around 0–15%) undergoes O-demethylation to morphine in the liver via CYP2D6
[17]
. However, because of the polymorphic nature of CYP2D6, the combination of alleles present can determine the extent to which this occurs. Importantly, the CYP2D6 ultra-rapid metaboliser phenotype (where there are more than two copies of the gene) has been associated with fatal cases of opiate toxicity in patients administered codeine, resulting from increased codeine biotransformation to morphine. This prompted the European Medicines Agency (EMA), the body that evaluates medicinal products for use in Europe, to review codeine use and led to the current restrictions by the Medicines and Healthcare Products Regulatory Agency (MHRA), the regulatory body for medicines and medical devices in the UK[18]
. Around 2% of Caucasians are ultra-rapid metabolisers, while 29% of Ethiopians carry more than two copies of the gene[15]
.
Identification of poor metabolisers is also important to circumvent issues such as clopidogrel resistance. Like codeine, clopidogrel is a prodrug that requires biotransformation into the active thiol group-containing moiety that inhibits ADP-induced platelet activation. The degree of platelet inhibition among individuals taking clopidogrel is highly variable[19]
, and this can potentially result in life-threatening sequelae such as stent thrombosis. CYP2C19 is one of the principal enzymes involved in clopidogrel activation and around 3–5% of Caucasians are deemed poor metabolisers, rising to 12% to 23% of the Asian population[19]
[20]
. The Food and Drug Administration (FDA), the body responsible for evaluating the safety and efficacy of medicines in the United States, issued a black box warning in 2010 advising about this association[21]
.
Transporter genes
In addition to determining enzyme function, polymorphisms also affect drug transporter proteins, structures responsible for the movement of ions and molecules across biological membranes. SLCO1B1, a gene encoding a protein known as OATP1B1, responsible for the intrahepatic transport of multiple compounds, including statins. Some SLCO1B1 variants have been associated with lower uptake of statins (in particular, simvastatin), increased drug exposure, and a higher risk of simvastatin intolerance and myopathy[22]
.
HLA genes
Pharmacogenomics facilitates insight into the underlying mechanisms of ADRs and can predict susceptibility to some idiosyncratic (type B) ADRs. An important class of genes are the human leukocyte antigens (HLA); specific HLA alleles can increase susceptibility to immune-mediated idiosyncratic ADRs through off-target mechanisms. An example is the effect of HLA-B*57:01 on abacavir hypersensitivity[23]
. The summary of product characteristics (SPC) for abacavir mandates HLA-B*57:01 typing before the use of abacavir. This is also mentioned in guidelines from organisations, such as the British HIV Association, who recommend pre-emptive HLA genotyping to identify high-risk individuals[24]
. Another example is the association of HLA-B*15:02 with carbamazepine-induced toxic epidermal necrolysis[25]
; this HLA allele is particularly prevalent in Southeast Asian countries.
Improving patient outcomes is just one of the potential benefits of pharmacogenomics. In an environment of increasing demand and limited resources, the projected annual costs to the NHS of medication-related hospital admissions is around £466m[5]
with a further £300m spent on unused medications[5]
[26]
.
A personalised prescribing approach strives to reduce ADRs and ineffective prescriptions to provide a smarter, more cost-effective way to manage the medicines budget. However, implementation of such innovative approaches has many challenges.
Barriers to implementation
Recent decades have seen multiple advances within the field of genomics; not least the completion of the Human Genome Project in 2003[27]
. This milestone and the statement of requirement for a ‘genetically literate’ primary care workforce by the Human Genetics Commission[28]
set high expectations for a paradigm shift towards genomics becoming routine in clinical care, spearheaded by pharmacogenomics. However, nearly 14 years later, even the integration of pharmacogenomics into standard clinical practice overall has been limited.
Multiple factors have been cited as reasons for this slow transition. First, there is a lack of understanding of the topic and its application among healthcare professionals. It is a relatively novel discipline, and the knowledge base therefore remains incomplete. Second, a lack of evidence-based implementation guidelines is a large hindrance for the field’s appropriation into general clinical practice. Third, ethical issues surrounding genetic information may be of concern to some patients and healthcare professionals. Fourth, the lack of clear guidance on the requirements and consequences of testing continue to deter patients and healthcare professionals alike[29]
.
Another major obstacle to implementation is the availability and expense of testing. However, the cost is decreasing all the time: the first human genome cost in excess of US$3bn; the cost now is <$1000 with the advent of next-generation sequencing technologies[30]
. However, equipment, trained personnel and strict quality control procedures are still required, and seldom available outside specialist units.
Finally, deficits in prescriber knowledge surrounding translating genetic information into clinical action points[31]
may also contribute to the poor uptake of testing. Ongoing efforts to collate evidence and information for clinical utilisation have been fruitful and the Pharmacogenomics Knowledgebase provides a repository of pharmacogenomic-based drug dosing clinical guidelines from multiple sources including the Clinical Pharmacogenetics Implementation Consortium (CPIC) and Dutch Pharmacogenetics Working Group (DPWG)[32]
.
Despite these barriers, there are some genomic tests within the UK that have become fundamental to routine care. For example, thiopurine S-methyltransferase (TMPT) polymorphisms, which can account for around 10% of overall thiopurine toxicity[33]
. As a result, the British National Formulary now recommends phenotype or genotype screening as standard before initiation.
Moving to the future
Despite the challenges, the future remains optimistic for pharmacogenomics. The Translational Pharmacogenetics Program is an implementation initiative instigated by the US Pharmacogenomics Research Network (PGRN) and aims to overcome the barriers that currently limit pharmacogenomic testing in clinical practice[34]
.
Simultaneously, within Europe, the Ubiquitous Pharmacogenomics Consortium (U-PGx) is commencing the first large, international implementation project to determine the impact of pre-emptive testing on ADR frequency, severity and associated costs[35]
.
Industry and regulatory bodies have begun to adapt, spurred by opportunities to re-trial previously abandoned compounds (e.g. bucindolol) or reduce ADRs in marketed products. As a result, incorporating genomic data into drug design and safety information is becoming common practice. An analysis of 517 EMA SPCs documented that nearly 15% contained utilisable pharmacogenomic information[36]
.
This shift into precision medicine is changing the business model of the pharmaceutical industry, moving away from the generic ‘block-buster’ style of drug development into, potentially more expensive, receptor or biomarker targeted therapies such as imatinib or ivacaftor[37]
.
This transition has forced the industry to view its role beyond solely the manufacture of the therapeutic and into the realms of companion diagnostics. These kits and assays allow precision medicine to be truly effective by identifying, as accurately as possible, the patient’s underlying disease mechanism or predictive biomarkers. Such technologies are already being used in clinical practice, with regulatory agencies approving cetuximab to be used as part of the first-line regime for KRAS mutation-negative (wild-type) Epidermal Growth Factor Receptor (EGFR)-expressing metastatic colorectal cancer as well as approving the first KRAS companion diagnostic test kit, the QIAGEN Therascreen[38]
.
It is important to recognise, however, that while the germline genome remains constant over a lifetime, gene expression levels may change. The realisation of personalised medicine will require holistic assessment of the patient that will include not only genomics, but also environmental factors, disease status and age-related changes.
In the UK, the NHS outlined its plans for integration of personalised medicine in 2016, aiming for whole-genome sequencing to be standard for specific conditions by 2020[39]
. The advent of the 100,000 Genomes Project[40]
and the sale from retail pharmacies of the home genetics kit, 23andMe®
[41]
, are further indicators of the need for pharmacists, in all areas of the profession, to become familiar with the principles of pharmacogenomics and its evolving application into clinical practice.
Financial and conflicts of interest disclosure:
The authors have no relevant affiliations or financial involvement with any organisation or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript. No writing assistance was used in the production of this manuscript.
Reading this article counts towards your CPD
You can use the following forms to record your learning and action points from this article from Pharmaceutical Journal Publications.
Your CPD module results are stored against your account here at The Pharmaceutical Journal. You must be registered and logged into the site to do this. To review your module results, go to the ‘My Account’ tab and then ‘My CPD’.
Any training, learning or development activities that you undertake for CPD can also be recorded as evidence as part of your RPS Faculty practice-based portfolio when preparing for Faculty membership. To start your RPS Faculty journey today, access the portfolio and tools at www.rpharms.com/Faculty
If your learning was planned in advance, please click:
If your learning was spontaneous, please click:
References
[1] Vogel F. Moderne problem der humangenetik. Ergeb Inn Med U Kinderheilk 1959;12:52–125. doi: 10.1007/978-3-642-94744-5_2
[2] Newbert DW. Pharmacogenetics and pharmacogenomics: why is this relevant to the clinical geneticist? Clin Genet 1999;56(4):247–258. doi: 10.1034/j.1399-0004.1999.560401.x
[3] Marshall A. Genset-Abbott deal heralds pharmacogenomics era. Nat Biotechnol 1997;15:829–830. doi: 10.1038/nbt0997-829b
[4] Spear BB, Heath-Chiozzi M, Huff J. Clinical application of pharmacogenetics. Trends Mol Med 2001;7(5):201–204. doi: 10.1016/S1471-4914(01)01986-4
[5] Pirmohamed M, James S, Meakin S et al. Adverse drug reactions as cause of admission to hospital: prospective analysis of 18 820 patients. BMJ 2004;329:15–19. doi: 10.1136/bmj.329.7456.15
[6] Wei MY, Ito MK, Cohen JD et al. Predictors of statin adherence, switching, and discontinuation in the USAGE survey: understanding the use of statins in America and gaps in patient education. J Clin Lipidol 2013;7(5):472–483. doi: 10.1016/j.jacl.2013.03.001
[7] Berger SL, Kouzarides T, Shiekhattar R et al. An operational definition of epigenetics. Genes Dev 2009;23(7):781–783. doi: 10.1101/gad.1787609
[8] Dunnenberger HM, Crews KR, Hoffman JM et al. Preemptive clinical pharmacogenetics implementation: current programs in five US medical centers. Annu Rev Pharmacol Toxicol 2015;55:89–106. doi: 10.1146/annurev-pharmtox-010814-124835
[9] Ritter J, Lewis L, Mant T et al. A Textbook of Clinical Pharmacology and Therapeutics, Fifth ed. London, UK: Hodder Arnold; 2008.
[10] Ravnan MC & Matalka MS. Vemurafenib in patients with BRAF V600E mutation–positive advanced melanoma. Clin Ther 2012;34(7):1474–1486. doi: 10.1016/j.clinthera.2012.06.009
[11] National Institute of Clinical Excellence: Technology appraisal guidance [TA107]: Trastuzumab for the adjuvant treatment of early-stage HER2-positive breast cancer (2006). Available at: https://www.nice.org.uk/guidance/ta107 (accessed October 2017)
[12] Condren ME & Bradshaw MD. Ivacaftor: a novel gene-based therapeutic approach for cystic fibrosis. J Pediatr Pharmacol Ther 2013;18(1):8–13. doi: 10.5863/1551-6776-18.1.8
[13] Meyer UA. Pharmacogenetics — five decades of therapeutic lessons from genetic diversity. Nat Rev Genet 2004;5:669–676. doi: 10.1038/nrg1428
[14] CYP2D6 allele nomenclature 2017. Available at: http://www.cypalleles.ki.se/cyp2d6.htm (accessed October 2017)
[15] Kalow W, Otton SV, Kadar D et al. Ethnic difference in drug metabolism: debrisoquine 4-hydroxylation in Caucasians and Orientals. Can J Physiol Pharmacol 1980;58(9):1142–1144. PMID: 7459706
[16] Zhou SF. Polymorphism of human cytochrome P450 2D6 and its clinical significance: Part I. Clin Pharmacokinet 2009;48(12):689–723. doi: 10.2165/11318030-000000000-00000
[17] Peck T & Hill S. Pharmacology for anaesthesia and intensive care, 4th ed. United Kingdom: Cambridge University Press; 2014.
[18] Medicines and Healthcare products Regulatory Agency. Codeine for analgesia: restricted use in children because of reports of morphine toxicity. Drug Safety Update 2013. Available at: https://www.gov.uk/drug-safety-update/codeine-for-analgesia-restricted-use-in-children-because-of-reports-of-morphine-toxicity (accessed October 2017)
[19] Serebruany V, Steinhubl SR, Berger PB et al. Variability in platelet responsiveness to clopidogrel among 544 individuals. J Am Coll Cardiol 2005;45(2):246–251. doi: 10.1016/j.jacc.2004.09.067
[20] Desta Z, Zhao X, Shin JG et al. Clinical significance of the cytochrome P450 2C19 genetic polymorphism. Clin Pharmacokinet 2002;41(12):913–958. doi: 10.2165/00003088-200241120-00002
[21] U.S. Food and Drug Administration. FDA Drug Safety Communication: Reduced effectiveness of Plavix (clopidogrel) in patients who are poor metabolizers of the drug, Postmarket Drug Safety Information for Patients and Providers 2010. Available at: https://www.fda.gov/Drugs/DrugSafety/PostmarketDrugSafetyInformationforPatientsandProviders/ucm203888.htm (accessed October 2017)
[22] SEARCH Collaborative Group: Link E, Parish S, Armitage J et al. SLCO1B1 variants and statin-induced myopathy — a genome-wide study. New Engl J Med 2008;359(8):789–799. doi: 10.1056/NEJMoa0801936
[23] Alfirevic A & Pirmohamed M. Drug induced hypersensitivity and the HLA complex. Pharmaceuticals (Basel) 2011;4(1):69–90. doi: 10.3390/ph4010069
[24] British HIV Association. Guidelines for the treatment of HIV-1-positive adults with ART. 2015 (2016 interim update). Available at: http://www.bhiva.org/documents/Guidelines/Treatment/2016/treatment-guidelines-2016-interim-update.pdf (accessed October 2017)
[25] Brent Ferrel P Jr & McLeod HL. Carbamazepine, HLA-B*1502 and risk of Stevens–Johnson syndrome and toxic epidermal necrolysis: US FDA recommendations. Pharmacogenomics 2008;9(10):1543–1546. doi: 10.2217/14622416.9.10.1543
[26] Department of Health. Pharmaceutical waste reduction in the NHS: a best practice compilation paper; 2015. Available at: https://www.england.nhs.uk/wp-content/uploads/2015/06/pharmaceutical-waste-reduction.pdf (accessed October 2017)
[27] National Human Genome Research Institute. All About The Human Genome Project (HGP). Available at: https://www.genome.gov/10001772/all-about-the–human-genome-project-hgp/ (accessed October 2017)
[28] Human Genetics Commission. Genes direct: ensuring the effective oversight of genetic tests supplied directly to the public 2003. Available at: http://www.conversations.canterbury.ac.nz/PHASEONE/resources/genetics/genes-direct.pdf (accessed October 2017)
[29] Kapoor R, Tan-Koi WC, Teo YY. Role of pharmacogenetics in public health and clinical health care: a SWOT analysis. Eur J Hum Genet 2016;24(12):1651–1657. doi: 10.1038/ejhg.2016.114
[30] Goodwin S, McPherson JD & McCombie WR. Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 2016;17(6):333–351. doi: 10.1038/nrg.2016.49
[31] Haga SB, Burke W, Ginsburg GS et al. Primary care physicians’ knowledge of and experience with pharmacogenetic testing. Clin Genet 2012;82(4):388–394. doi: 10.1111/j.1399-0004.2012.01908.x
[32] Swen JJ, Nijenhuis M, de Boer A et al. Pharmacogenetics: from bench to byte—an update of guidelines. Clin Pharmacol Ther 2011;89(5):662–673. doi: 10.1038/clpt.2011.34
[33] Ansari A, Arenas M, Greenfield SM et al. Prospective evaluation of the pharmacogenetics of azathioprine in the treatment of inflammatory bowel disease. Aliment Pharmacol Ther 2008;28:973–983. doi: 10.1111/j.1365-2036.2008.03788.x
[34] Shuldiner AR, Relling MV, Peterson JF et al. The Pharmacogenomics Research Network Translational Pharmacogenetics Program: overcoming challenges of real-world implementation. Clin Pharmacol Ther 2013;94:207–210. doi: 10.1038/clpt.2013.59
[35] van de Wouden CH, Cambon-Thomsen A, Cecchin E et al.; Ubiquitous Pharmacogenomics Consortium. Implementing Pharmacogenomics in Europe: Design and Implementation Strategy of the Ubiquitous Pharmacogenomics Consortium. Clin Pharmacol Ther 2017;101(3):341–358. doi: 10.1002/cpt.602
[36] Ehmann F, Caneva L, Prasad K et al. Pharmacogenomic information in drug labels: European Medicines Agency perspective. Pharmacogenomics J 2015;15(3):201–210. doi: 10.1038/tpj.2014.86
[37] Collier R. Bye, bye blockbusters, hello niche busters. CMAJ 2011;183(11):E697–E698. doi: 10.1503/cmaj.109-3874
[38] Eli Lilly. FDA Approves ERBITUX(R) (cetuximab) as First-Line Treatment in KRAS Mutation-Negative (Wild-Type) Epidermal Growth Factor Receptor (EGFR)-Expressing Metastatic Colorectal Cancer in Combination with FOLFIRI (Irinotecan, 5-Fluorouracil, Leucovorin). Available at: https://investor.lilly.com/releasedetail.cfm?releaseid=689686 (accessed October 2017)
[39] NHS England. Improving outcomes through personalised medicine. Available at: https://www.england.nhs.uk/wp-content/uploads/2016/09/improving-outcomes-personalised-medicine.pdf (accessed October 2017)
[40] Genomics England. The 100,000 Genomes Project. Available at: https://www.genomicsengland.co.uk/the-100000-genomes-project/ (accessed October 2017)
[41] Andalo D. Superdrug is first retailer in the world to sell DNA testing kit in store. Pharm J 2015;294(7858). doi: 10.1211/PJ.2015.20068268