In a recent BBC/ICM poll, the most common concern about hospital care was the risk of acquiring an infection, such as meticillin-resistant Staphylococcus aureus or Clostridium difficile, with 40 per cent of people surveyed listing the risk of infection as their biggest fear.[1]
In the fight against the spread of both nosocomial and community-acquired infections, attempts have been made to increase public awareness about the link between health and hand hygiene through a number of television, radio and advertising campaigns, including the “Hand in hand” campaign[2]
launched by the East Midlands Strategic Health Authority, which the charity MRSA Action UK suggested should be rolled out nationally.
Multidrug-resistant bacteria are indeed an increasing clinical problem,[3]
with a particular concern being the transmission of resistance between these bacteria, which could ultimately lead to strains that have limited or no susceptibility to antibacterial agents. For example, although the incidence of glycopeptide-resistant enterococci (GRE) in Europe is currently much lower than in the US (where more than 20 per cent of enterococcal isolates are vancomycin-resistant), a report in 2003 of an in vivo transmission of vancomycin resistance from GRE to MRSA highlights the significant risk that would be associated with having co-existing, non-isolated infections because of these pathogens.[4]
It should also be noted that MRSA is susceptible to very few agents, including the glycopeptides (vancomycin and teicoplanin), quinupristin-dalfopristin and linezolid. Cases of meticillin- and quinuprustin-dalfopristin-resistant S aureus have already been reported in Europe.[5]
From April 2009, the Care Quality Commission (CQC) took over responsibility for health and social care regulation from the Healthcare Commission. “The Health and Social Care Act 2008: code of practice for the NHS on the prevention and control of healthcare associated infections and related guidance” (the “hygiene code”), published in January 2009, describes how the CQC will assess compliance with the requirements regarding healthcare-associated infections, as set out in the regulations made under section 20(5) of this Act.[6]
Relevant NHS bodies must have, and adhere to, policies for the control of outbreaks and infections associated with both MRSA and C difficile, while acute NHS trusts must have similar policies for other specific alert organisms (see Panel 1).
Panel 1: Specific alert organism policies required by acute NHS trusts
The organisms include
- Meticillin-resistant Staphylococcus aureus
- Clostridium difficile
- Glycopeptide-resistant enterococci
- Acinetobacter and other antibiotic-related bacteria
- Tuberculosis (including multidrug-resistant TB)
- Respiratory viruses
- Diarrhoeal infections
- Viral haemorrhagic fevers
- Legionella
With specific regard to MRSA, this policy should make provision for the screening of all patients on admission, including the screening of all elective admissions since March 2009, and the provision for screening of emergency admissions on presentation as soon as practicable. This screening should then be used to inform the need for decontamination or isolation of colonised patients.
The Health Protection Agency publishes data derived from the mandatory surveillance of MRSA, C difficile and vancomycin-resistant enterococci (VRE) bacteraemia, and the most recent data show that, between January and March 2009, there was a 2.1 per cent increase in MRSA bacteraemia compared with the previous quarter (but a reduction of 29 per cent compared with the corresponding quarter in 2008), while there was a 34 per cent decrease in the number of reported MRSA bacteraemias in the financial year 2008–09.[7]
The need for rapid and simple methods for the detection of pathogenic bacteria, such as MRSA, GRE, extended-spectrum beta-lactamase-producing organisms (ESBL), Pseudomonas aeruginosa, and Acinetobacter baumannii, is thus self-evident. We will now concentrate on some techniques available for use in bacteraemia surveillance.
Techniques
Microscopy
Traditionally, pathogenic bacteria are detected by microscopy on the basis of their colonial appearance after inoculation of a culture medium that facilitates the growth of a wide range of organisms. Such identification requires the skills of an experienced clinical microbiologist and often requires further testing of commensal bacteria, which may have morphological characteristics similar to those of pathogenic bacteria.
Chromogenic detection
Specific chromogenic media,[8]
in which a non-coloured enzyme substrate (targeting molecule linked to chromogenic compound) is added to the culture medium, have been used for over 20 years in the detection of pathogenic bacteria (Figure 1). Ideally, this is a substrate for an enzyme that is unique to a particular bacterium (although this is rare) and cleavage of a key bond liberates a chromogen, which can be detected against a background of other colourless colonies (since these do not contain the requisite enzyme for cleavage of the chromogenic substrate). Often, more than one chromogenic substrate can be used to help in the differentiation of commensal and pathogenic bacteria.
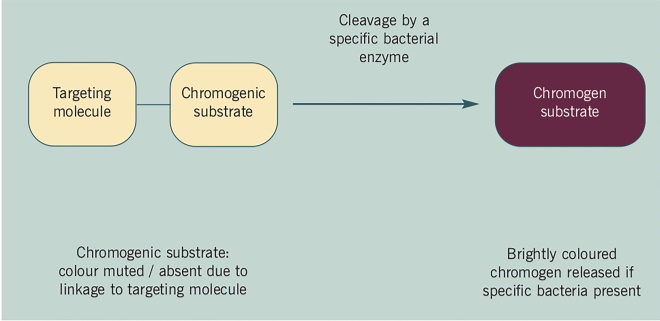
Figure 1: Schematic for chromogenic detection of specific bacteria
Among the benefits of the use of these media are that they can be sufficiently specific so that no further testing is required, and can give some indicative colours for bacterial colonies. However, the time required (usually 24–48 hours) for the growth of the colonies (and thus the development of colour) is currently a limiting factor in the development of a rapid test that could be applied to all patients on admission. Such chromogenic media have been used for the detection of MRSA-, GRE- and ESBL-producing organisms.
A recent example of a medium that is selective for the detection of P aeruginosa, the most common respiratory pathogen in patients with cystic fibrosis, was developed as part of our collaboration with the University of Northumbria and bioMeÌrieux, Marcy l’Etoile, France (Figure 2). It uses the pale yellow β-alanyl-1-pentylre- sorufamine, which is specifically cleaved by P aeruginosa β-alanyl aminopeptidase to give purple colonies with a metallic sheen that are easily detected by the naked eye.[9]
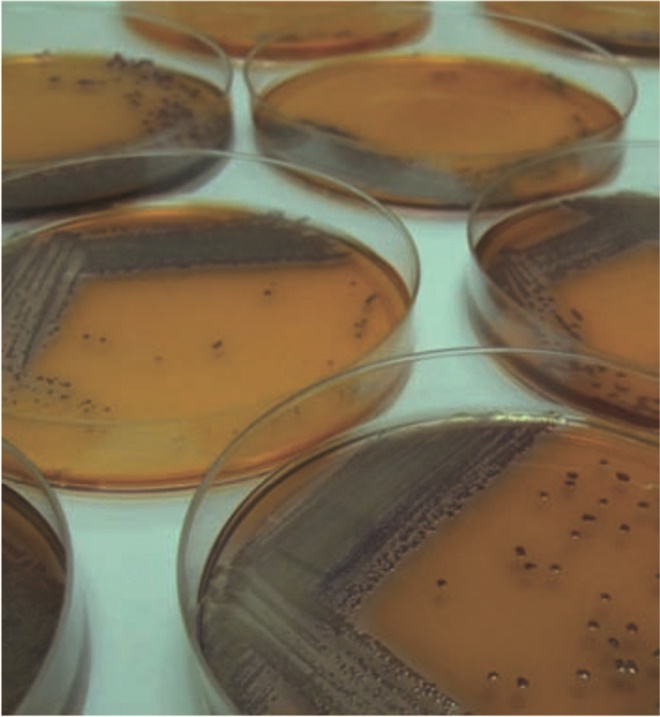
Figure 2: Detection of Pseudomonas aeruginosa by the specific hydrolysis of β- alanyl-1-pentylresorufamine by β-alanyl aminopeptidase and the origin of the purple colour (1-pentylresorufamine)
Fluorogenic media reduce the time required for such tests because the detection of fluorescence against a non-fluorescent background is more sensitive than the detection of a coloured compound against a lightly coloured background.
Automated laboratory systems based on chromogenic or fluorogenic substrates are available for bacterial identification, but these still require relatively lengthy sample turnaround times (usually 24–48 hours).
Molecular diagnostic methods
The advantages of molecular diagnostic methods are that: they are rapid (results can typically be obtained within a few hours), they can be highly specific and, like the automated culture-based methods, they can be performed in closed systems and have the capacity for automation. These methods are based on the bacterial genotype and often involve real-time polymerase chain reaction (PCR) technology.[10]
Conclusion
The increased requirement for bacterial surveillance techniques that are simple, rapid, cost-effective and specific means that the development of such tests is an area of continued research interest. Fully automated methods that are able to give results in hours rather than days are ideal since these would enable the rapid isolation and decontamination of colonised patients, resulting in a reduction in the opportunities for the spread of infections and cross-infection of patients by a number of multidrug-resistant bacteria.
The rapid and specific identification of pathogenic bacteria will also enable a patient’s treatment regimen to be specifically tailored to suit clinical need. This process will improve patient care and also help to combat the threat of future antibacterial resistance.
To see the original article, download the attached PDF (190K)
About the authors
Paul W Groundwater is professor of medicinal chemistry at the faculty of pharmacy at the University of Sydney, Australia.
Adam Todd, MRPharmS, is senior lecturer in pharmacy practice, Alan J Worsley, MRPharmS, is principal lecturer in pharmacy practice and Roz J Anderson is professor of pharmaceutical chemistry, all at Sunderland Pharmacy School, University of Sunderland.
References
[1] BBC News. Public tolerant of “nanny state”. Available at news.bbc.co.uk (accessed 19 August 2009).
[2] Hand in Hand: fighting infection together. Available at www.eastmidlands.nhs.uk (accessed 19 August 2009).
[3] Wickens H, Wade P. Understanding antibiotic resistance.The Pharmaceutical Journal 2005;274:501–4.
[4] Chang S, Sievert DM, Hageman JC, Boulton ML, Tenover FC, Pouch Downes F et al. Infection with vancomycin-resistant Staphylococcus aureus containing the vanA resistant gene. New England Journal of Medicine 2003;348:1342–7.
[5] Werner G, Cuny C, Schmitz F-J, Witte W. Methicillin-resistant, quinuprustin-dalfopristin-resistant Staphylococcus aureus with reduced sensitivity to glycopeptides. Journal of Clinical Microbiology 2001;39:3586–90.
[6] The Health Act 2006: code of practice for the prevention and control of healthcare associated infections. Available at www.dh.gov.uk (accessed 19 August 2009).
[7] Summary points on quarterly (April 2006 to March 2009) and financial year (April 2006 to March 2009) MRSA bacteraemia data derived from mandatory surveillance, June 2009. Available at www.hpa.org.uk (accessed 19 August 2009).
[8] Perry JD, FreydieÌ€re AM. The application of chromogenic media in clinical microbiology. Journal of Applied Microbiology 2007;103:2046–55.
[9] Zaytsev AV, Anderson RJ, Bedernjak A, Groundwater PW, Huang Y, Perry JD et al. Synthesis and testing of chromogenic phenoxazinone substrates for β-alanyl aminopeptidase. Organic and Biomolecular Chemistry 2008;8:682–92.
[10] Tenover FC. Rapid detection and identification of bacterial pathogens using novel molecular technologies: infection control and beyond. Clinical Infectious Diseases 2007:44;418–23.


