James King-Holmes / Alamy Stock Photo
After reading this article, you should be able to:
- Explain what DPYD genetic testing is, as well as how and why it is used;
- Understand the relevance of this testing to pharmacists;
- Envision how pharmacogenomics could become part of a routine patient care pathway.
Pharmacogenomics, also referred to as PGx, is the study of how the human genome influences the way the body processes and responds to medicines[1]. Although there are many well-characterised examples of drug–gene relationships, few pharmacogenetic tests have been adopted into routine clinical practice in the NHS. A 2022 report by the Royal College of Physicians and the British Pharmacological Society highlighted the potential for pharmacogenomic testing to be implemented on a far broader scale, to increase the likelihood that individuals receive the most effective medicine the first time they are treated at the most appropriate dose[2].
In the UK, it is estimated that 3–6% of the population have absolute or partial deficiency of the dihydropyrimidine dehydrogenase (DPD) enzyme[3]. This deficiency can result in risk of severe or even fatal toxicity from adverse reactions to certain curative and palliative cancer treatments[3]. Systemic fluoropyrimidine chemotherapy agents (5-fluorouracil [5-FU] and the prodrugs capecitabine and tegafur) are commonly used for a range of different cancer types and, although they are generally well tolerated, severe drug reactions (such as neutropenia, diarrhoea and mucositis) occur in 5–10% of the treated population[3].
When metabolising 5-FU, the rate-limiting step is the DPD enzyme rendering the chemotherapy agent inactive so a DPD deficiency can greatly increase the severity of the reaction[4]. Genetic variants in the DPYD gene have been associated with DPD deficiency, meaning that pre-emptive genetic testing is an effective means of reducing harm to patients. DPYD genetic screening is now mandated in England before the administration of systemic fluoropyrimidine-based chemotherapy, making this one of the first pharmacogenomic tests to be applied nationally[5].
This article will provide an overview of DPYD genetic testing, explain how and why it is used in practice and its relevance to pharmacy teams. It will also show how DPYD testing can potentially become an exemplar pathway for the implementation of pharmacogenomics in the NHS.
DPYD pharmacogenomic testing
All genomic tests commissioned by NHS England and NHS Improvement are available through the National Genomic Test Directory[6]. The directory specifies which genomic tests are commissioned, the technology through which they are available and the patients who are eligible for testing. For cancer patients eligible for DPYD testing, a blood sample is taken and sent to one of seven Genomic Laboratory Hubs (GLHs) in England, where the test is performed and reported (see Box 1)[6].
Box 1: Genomic Laboratory Hubs in England
- Central and South Genomic Laboratory Hub, led by Birmingham Women’s and Children NHS Foundation Trust;
- East Genomic Laboratory Hub, led by Cambridge University Hospitals NHS Foundation Trust;
- North West Genomic Laboratory Hub, led by Manchester University NHS Foundation Trust;
- North Thames Genomic Laboratory Hub, led by Great Ormond Street Hospital for Children NHS Foundation Trust;
- South East Genomic Laboratory Hub, led by Guy’s and St Thomas’ NHS Foundation Trust;
- South West Genomic Laboratory Hub, led by North Bristol NHS Trust;
- North East and Yorkshire Genomic Laboratory Hub, led by The Newcastle upon Tyne Hospitals NHS Foundation Trust.
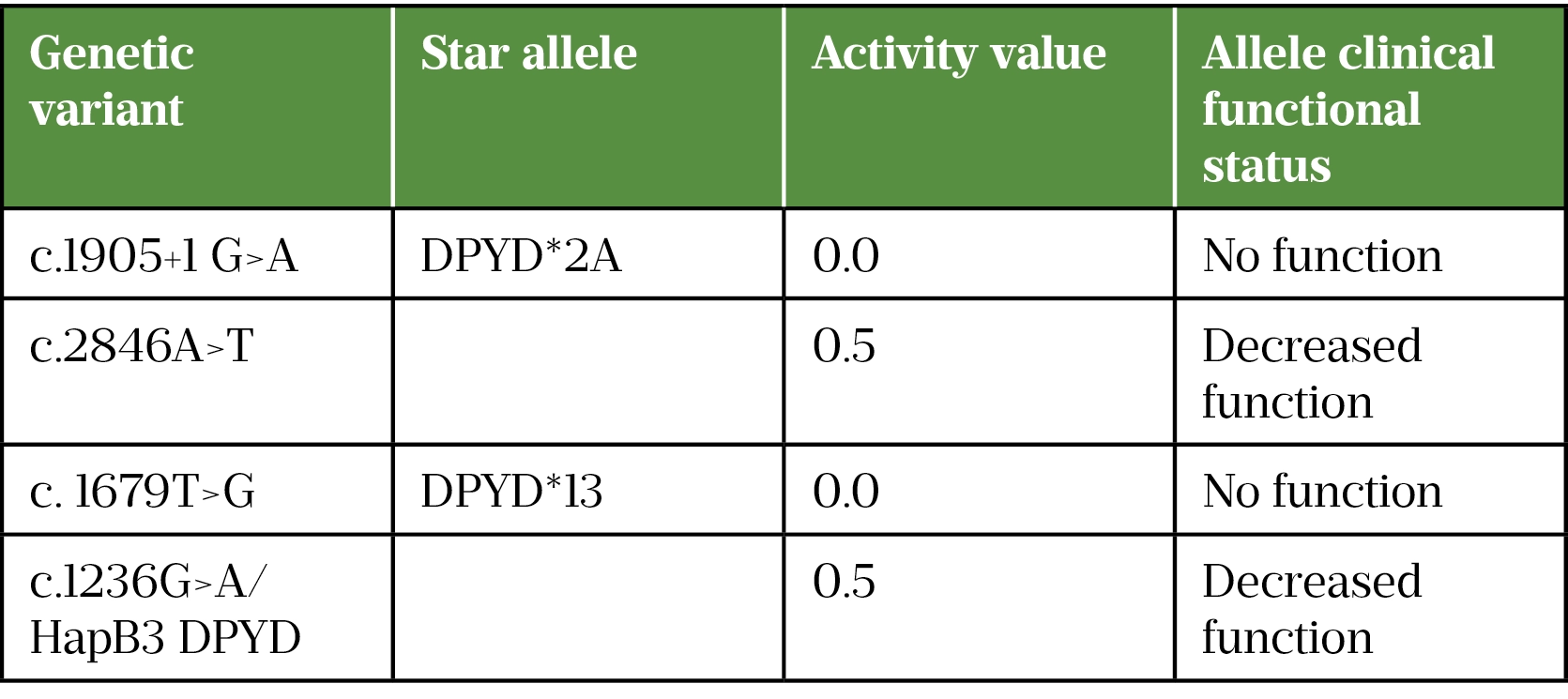
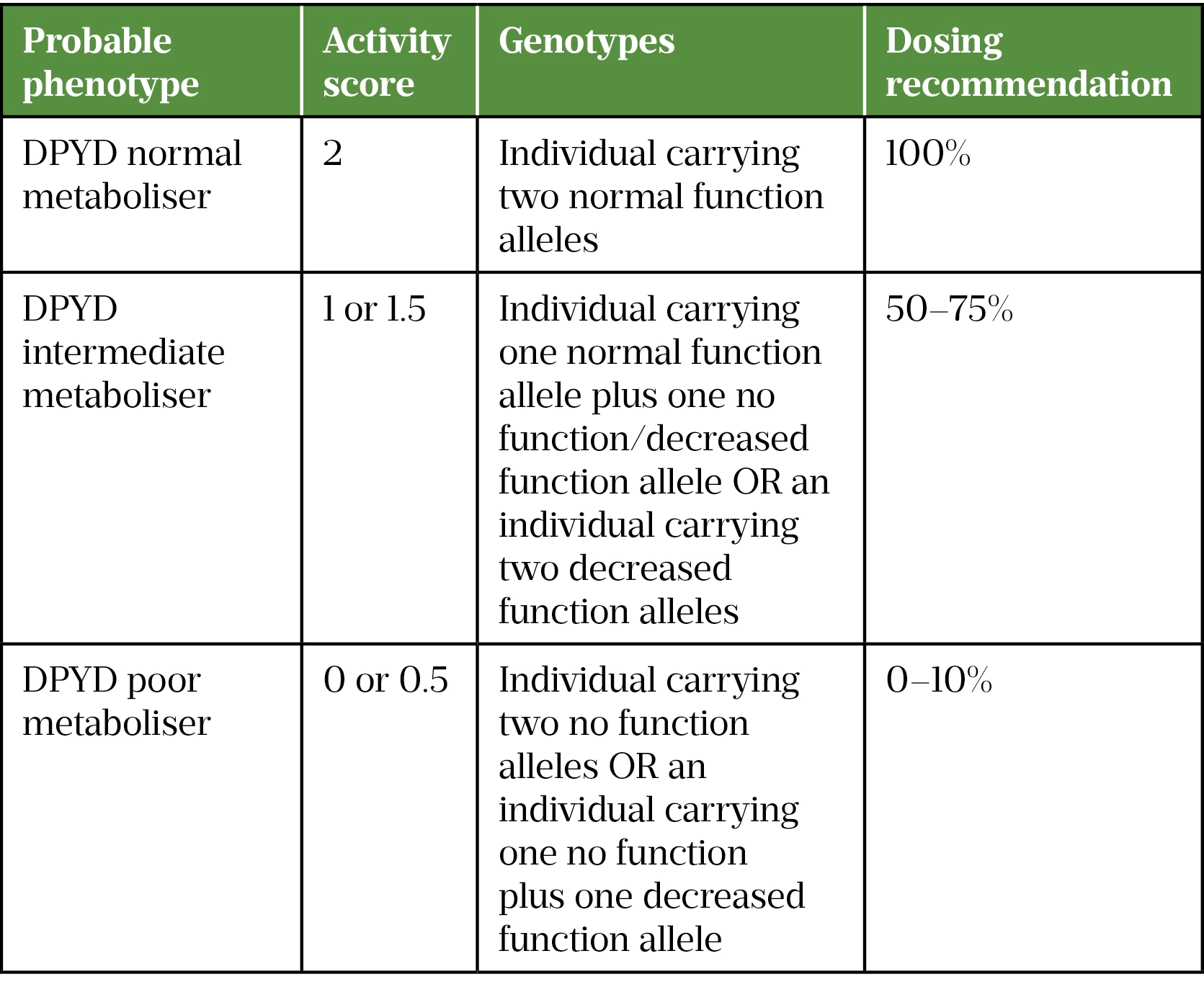
The DPYD pharmacogenomic test sits within the National Genomic Test Directory for Cancer and is carried out using a ‘hotspot’ test. Rather than looking for variants across the whole gene, the test is targeted to detect four specific variants. These variants have been chosen as the most clinically relevant because of their population frequency, established impact on DPD function and risk of toxicity[7]. See Table 1 for the four genetic variants targeted in DPYD testing[7,8].
Pharmacogenomic nomenclature
The nomenclature used in pharmacogenetics can seem complex but is relatively easy to understand once you apply it to specific examples. The nomenclature for the DPYD variant is shown in Figure 1.
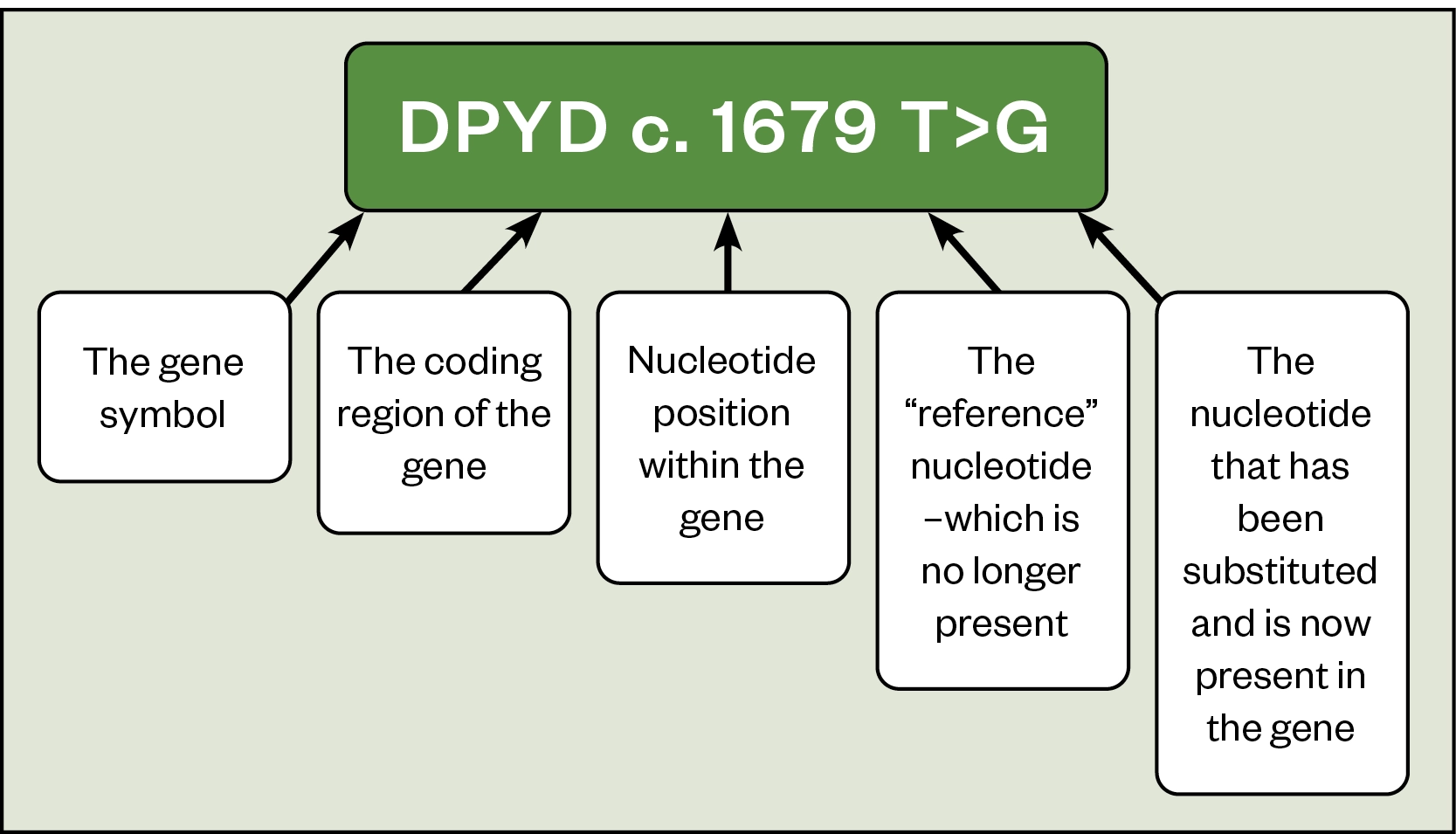
The c in c.1679T>G indicates that the change is within the coding region of the DPYD gene, followed by the number to represent the position of the change at 1,679 (i.e. the 1,679th nucleotide in the gene). T>G indicates that a thymine (T) has been replaced by a guanine (G). In this example a single nucleotide in a single position has been substituted and this genetic change is known as a single nucleotide polymorphism (SNP). It represents the most common type of genetic change[8].
Since the raw variant data demonstrated in Figure 1 is too detailed for the needs of most healthcare professionals, simpler star (*) allele names are given to raw genetic variants. In star-allele nomenclature, *1 is the first designated reference sequence to which polymorphic sites are then compared. This is usually the first sequence that was described to code for the functional protein product and may not be the most common allele in every ethnic population[9]. A unique number (e.g. DPYD*2A) is then given to each novel variant. Not all DPYD variants have been assigned a star allele, so the preferred nomenclature for DPYD variants is the Human Genome Variation Society or rsID number[7]. Depending on which version of the human genome is used as reference will change the rsID that the variant is compared to. Full discussion of this is beyond the scope of this article but further detail can be found here.
For some pharmacogenes, including DPYD, genotype–phenotype relationships have been characterised sufficiently well for the variants considered that it is possible to assign a gene activity score ‘AS’ to them[7]. This makes it possible to produce dosing guidelines that account for the range of genetic variation that may be detected by the DPYD hotspot test[3,7].
The patient pathway
Since the human genome is made up of two sets of chromosomes, one inherited from each parent, there are usually two copies of each gene in every cell in the body. It is therefore possible to have a variant on one copy of the gene and not the other. As a result, patients who are found to have a genetic variant can be considered:
- Heterozygous (a variant on one copy of the gene);
- Homozygous (the same variant on both copies of the gene);
- Compound heterozygous (two different variants, one on each copy of the gene).
The activity score is measured for both alleles and a combined sum is calculated. From this combined score, a probable phenotype/metaboliser status can be assigned to the patient.
Figure 2 shows an example patient pathway for DPYD testing. It highlights the importance of the pharmacist in reviewing the DPYD test result and working with the prescriber to enact dosage recommendations in the context of the individual patient’s prescription. Additionally, pharmacists need to ensure that the any dose modifications made on the basis of test results are reviewed and titrated based on patient tolerance (see Table 2[3,8]).
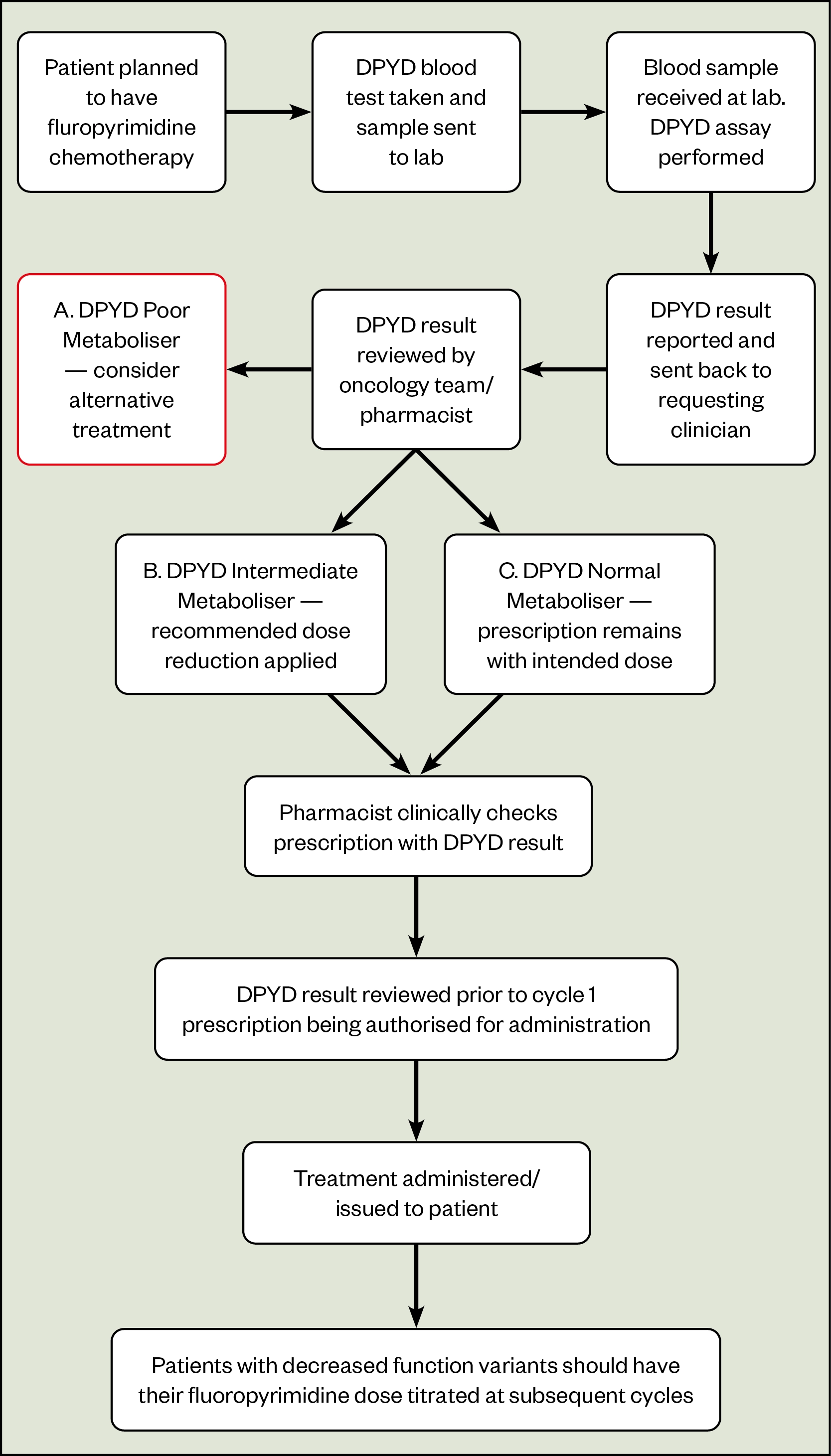
Adapted from guidance from the Clinical Pharmacogenetics Implementation Consortium and the UK Chemotherapy Board[3]
Patients who do not have any genetic variants are considered DPYD normal metabolisers (DPYD-AS: 2) and will be administered the initially intended dose of fluoropyrimidine-based chemotherapy[3]. Patients who have a variant identified on one copy of the gene (heterozygotes) are classed as DPYD intermediate metabolisers (DPYD-AS: 1 or 1.5) and should have their fluoropyrimidine dose reduced according to the specific variant identified.
Dose adjustments should be based on the advice given on the genetic report or other local or national guidance, such as the UK Chemotherapy Board national guidance (depending on the standard operating policy at the treatment provider)[3]. Patients may also have their dose modified on the basis of other factors, such as renal function and other comorbidities, since pharmacogenomics is just one aspect of medicines optimisation.
Patients identified as homozygous or compound-heterozygous for a reduced functioning DPYD variant are categorised as DPYD poor metabolisers (DPYD-AS: 0) and will generally be considered for alternative treatments because of the severe risk of toxicity with fluoropyrimidines. However, in the absence of suitable alternatives and as part of a shared decision-making process with the patient, consultant oncologists and pharmacists may decide to proceed with fluoropyrimidine-based therapy at heavily reduced doses[3]. This conversation would be specific to the individual patient’s diagnosis and available treatment options, the variant(s) identified and other patient-specific factors to enable a risk versus benefit judgement to be made.
Patient counselling
Counselling for DPYD testing does not require specialist genetic counselling services and is often undertaken at the time of consent for chemotherapy by an oncologist or non-medical prescriber. When discussing pharmacogenomic testing with patients, it is useful to provide the context in which the result will be used: to aid drug and/or dose selection according to guideline recommendations and to reduce the risk of severe or life-threatening side effects from fluoropyrimidine-based chemotherapy treatment. In this way, DPYD testing is not dissimilar to renal or liver function testing as it can guide prescribing decisions and optimise drug therapy[2].
Patient autonomy and informed consent are essential to the medical decision-making process. The consent process for pharmacogenomic testing is the same as for any medical procedure — patients and their carers must be provided with appropriate information to make informed decisions about their health. More detailed and technical information may also be required to ensure that patients get the right information for their needs. Written information and relevant online resources should also be offered to patients and carers. Box 2 lists the counselling points to discuss with patients.
Box 2: Counselling points
- DPYD testing is being offered to guide treatment and/or dose selection and to reduce the risk of severe or life-threatening side-effects from fluoropyrimidine chemotherapy treatment;
- If a clinically relevant variant is identified, changes will be made to the chemotherapy regime, such as an initial dose reduction. The dose can be increased subsequently if there are no serious side effects;
- If no clinically relevant toxicity-related variants are identified, there is still a risk of toxicity owing to other environmental, genetic or disease-related factors. Patients should be advised to report any side effects to the oncology unit without delay;
- Family members are not routinely tested when a patient is identified as carrying a clinically relevant variant, because a DPD deficiency does not usually cause symptoms unless fluorouracil or capecitabine chemotherapy is administered;
- Local procedures for the storage of blood samples should also be discussed with the patient;
- The DNA sample will not be used for other genetic tests without the patient’s consent.
The genetic report
Figure 3 shows the type of information included on a genetic report, including the rationale for testing, a summary of the results and technical information about the test itself. If dosing recommendations are provided, these should be considered as part of a holistic assessment of the patient.

The GLH will return a genetic report similar to the one seen in Figure 3 to the requesting clinician. The provider is then responsible for incorporating the patient’s DPYD genetic report into their management plan and communicating results to the patient. Based on the individual patient’s result and the standard operating procedures at the trust, oncology teams may adjust the dose as described in the patient pathway.
Targeted testing technology
Most of the genotyping is undertaken as per the requirements of the National Genomic Test Directory, using simple targeted testing technology performed on a standard blood sample. The advantage of the targeted gene testing approach is that result turnaround time is 24 to 72 hours (the current NHS mandate is less than five working days)[6]. This means the results can be delivered in a clinically relevant timeframe to inform treatment decisions in the patient pathway.
Future implications
The implementation of national DPYD screening is one of seven transformation projects delivered by the NHS Genomic Medicines Service Alliances (GMSA) in 2021/2022 that aim to embed genomics into routine patient care pathways[10]. The DPYD project is aimed to optimise and improve the clinical effectiveness of DPYD gene testing for patients with cancer across England. GMSA regional pharmacy leads have been able to feed back on the positive contribution of pharmacists in chemotherapy provider trusts around the country in embedding DPYD pharmacogenomic testing into routine oncology pathways.
The main messages identified by the project leads from the implementation of the DPYD pathway include:
- The importance of national commissioning to ensure testing is available across the country;
- The value of communication between clinical teams and regional GMSA leads;
- The importance of pharmacy teams in taking ownership/leadership of pharmacogenomic testing;
- The need for decision support tools/dosing guidelines (such as the UK Chemotherapy Board’s national dosing guidance[3]);
- The requirement for interoperability between electronic systems to permit reporting within ‘clinically relevant’ timeframes;
- That genomics is only one piece of a complex puzzle and that adverse drug reactions are influenced by other patient factors (e.g. comorbidities).
Future work aims to characterise less common DPYD variants in patients who experience severe toxicity despite no variants being detected on initial screening.
The future of pharmacogenomics
To support the integration of pharmacogenomics into patient care pathways, consideration must also be given to alternative and newly emerging testing approaches. For example, pre-emptive testing with pharmacogenomic gene panels can test multiple genes simultaneously, improving efficiency and cost-effectiveness when compared with multiple individual genetic tests[11]. Having genomic data readily available in the electronic patient record at the point of prescribing would eliminate concerns related to turnaround time in clinical scenarios where there is an urgent need to initiate treatment; for example, before the use of clopidogrel in percutaneous coronary intervention[12].
Of course, there are challenges to this approach, not least the need to prove cost-effectiveness in the selected population and a requirement for comprehensive clinical decision support tools[13]. If successfully incorporated into the electronic patient record, and with sufficient integration to permit healthcare providers across sectors to view and act upon a patient’s pharmacogenomic information, the potential benefits are enormous[2].
Cancer learning ‘hub’
Pharmacists are playing an increasingly important role in supporting patients with cancer, working within multidisciplinary teams and improving outcomes.
However, in a rapidly evolving field with numbers of new cancer medicines is increasing and the potential for adverse effects, it is now more important than ever for pharmacists to have a solid understanding of the principles of cancer biology, its diagnosis and approaches to treatment and prevention.
This new collection of cancer content, brought to you in partnership with BeOne Medicines, provides access to educational resources that support professional development for improved patient
- 1Pharmacogenomics. Genomics Education Programme. https://www.genomicseducation.hee.nhs.uk/glossary/pharmacogenomics (accessed Jun 2022).
- 2Personalised prescribing: using pharmacogenomics to improve patient outcomes. Royal College of Physicians. 2022.https://www.rcp.ac.uk/projects/outputs/personalised-prescribing-using-pharmacogenomics-improve-patient-outcomes (accessed Jun 2022).
- 3Personalised Medicine Approach For Fluoropyrimidine-based Therapies. UK Chemotherapy Board. 2020.https://www.theacp.org.uk/userfiles/file/resources/dpd-testing-ukcb-july-2020-updated.pdf (accessed Jun 2022).
- 4Thorn CF, Marsh S, Carrillo MW, et al. PharmGKB summary. Pharmacogenetics and Genomics. 2011;21:237–42. doi:10.1097/fpc.0b013e32833c6107
- 5Clinical Commissioning Urgent Policy Statement Pharmacogenomic testing for DPYD polymorphisms with fluoropyrimidine therapies [URN 1869] (200603P). NHS England. https://www.england.nhs.uk/wp-content/uploads/2020/11/1869-dpyd-policy-statement.pdf (accessed Jun 2022).
- 6National Genomic Test Directory. NHS England. 2018.https://www.england.nhs.uk/publication/national-genomic-test-directories (accessed Jun 2022).
- 7Amstutz U, Henricks LM, Offer SM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Dihydropyrimidine Dehydrogenase Genotype and Fluoropyrimidine Dosing: 2017 Update. Clin. Pharmacol. Ther. 2017;103:210–6. doi:10.1002/cpt.911
- 8Various types of variant: what is genomic variation? Genomics Education Programme. 2016.https://www.genomicseducation.hee.nhs.uk/blog/various-types-of-variant-what-is-genomic-variation (accessed Jun 2022).
- 9Robarge JD, Li L, Desta Z, et al. The Star-Allele Nomenclature: Retooling for Translational Genomics. Clin Pharmacol Ther. 2007;82:244–8. doi:10.1038/sj.clpt.6100284
- 10Our regional and national projects — 2021-2022 projects. North West Genomic Medicine Service Alliance. https://www.nw-gmsa.nhs.uk/about-us/our-regional-and-national-projects/2021-2022-projects (accessed Jun 2022).
- 11Turner RM, Newman WG, Bramon E, et al. Pharmacogenomics in the UK National Health Service: opportunities and challenges. Pharmacogenomics. 2020;21:1237–46. doi:10.2217/pgs-2020-0091
- 12Zhou Y, Armstead A, Coshatt G, et al. Comparison of Two Point-of-Care CYP2C19 Genotyping Assays for Genotype-Guided Antiplatelet Therapy. Ann Clin Lab Sci 2017;47:738–43.https://www.ncbi.nlm.nih.gov/pubmed/29263048
- 13Guidance for using pharmacogenomic information in clinical practice. Professional Record Standards Body . 2020.https://theprsb.org/wp-content/uploads/2020/11/Pharmacogenomics-Report_V1.0.pdf (accessed Jun 2022).


