Mclean/Shutterstock.com
Pharmacogenomics, a field combining pharmacology and the study of the genome, is set to revolutionise the way we treat our patients in the UK[1]. Where there is now an understanding that genetics is behind the cause of a disease or treatment response, we are becoming more able to tailor treatment to the individual in order to select the most effective treatment strategy, reduce the burden of adverse events and prescribe the ideal dose[1]. However, it will only bring equitable benefits to patients if research and clinical recommendations are reflective of our diverse population and we move our thinking away from racial categories to genetically-determined ancestries.
Race does not equate to biology
We all share 99.9% of our DNA, but the 0.1% which makes each of us unique can have a significant impact on our health, and the subsequent care we require[2,3]. Differences between people have been described in terms of race, ethnicity or ancestry, and all have been used interchangeably in scientific literature to group people together. In some papers, the terminology of white, black and Asian are used, and in others you will see reference to European, African or Asian ethnicity as an alternative.
However, race is a social construct which does not represent biology and does not capture the rich diversity within populations[2–5]. For example, there is more genetic diversity within the frequently used definition of African populations compared to Asian populations, and understanding this diversity is vital to ensure we apply pharmacogenomics correctly for individuals[6]. This within-population diversity can be exemplified by the gene test for the cytochrome P450 (CYP) enzyme, CYP2D6, which metabolises 25% of all drugs in clinical use[7]. The gene for CYP2D6 has more than 100 variants, and the greatest heterogeneity for this enzyme is found in the population from the African continent, compared to other continents. This population also shows the greatest genetic variation for other CYP enzymes[7–9].
We need to move away from thinking along the lines of racial and ethnic constructs within pharmacogenomics and focus on genetically-determined ancestry
There are several variants within the genes for CYP enzymes (pharmacogenes) that are functional and lead to a particular enzyme-metabolising trait, also known as a phenotype[10]. Examining the distribution of these variants, in which the variations are denoted with stars, show that CYP2D6*10 and CYP2D6*41 — which lead to poor drug metabolism — are more commonly present within Asian and European ancestry[10]. However, CYP2D6*17 and CYP2D6*29 variants, which also lead to poor drug metabolism, are common within African American ancestry[7]. If we look at CYP2D6 ultrarapid metabolisers, data indicate that 28% of North Africans have the variants that would class them as ultrarapid metabolisers compared to only 3% of African Americans, highlighting significant differences within a population, which we would not be able to decipher if individuals were categorised broadly as black[6]. Therefore, we need to move away from thinking along the lines of racial and ethnic constructs within pharmacogenomics and focus on genetically-determined ancestry to guide how we can better optimise prescribing in terms of drug safety and efficacy[4].
Lack of data
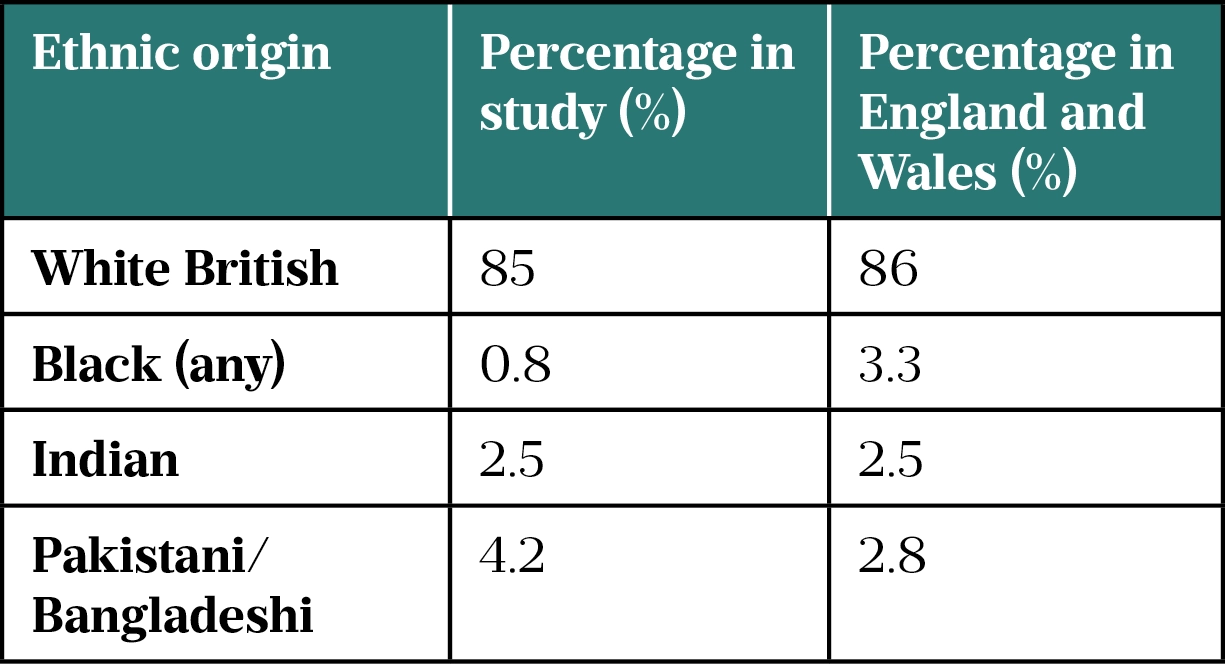
In 2021, it was reported that 86% of genomic studies have been conducted in people of European ancestry[11]. In the UK, participants in the 100,000 Genomes Project — a scheme to sequence genomes from around 85,000 NHS patients affected by a rare disease or cancer, run by Genomics England — are 85% white, with only 0.8% of participants identifying as black (see Table)[3,12].
This lack of representation within trials means that we cannot understand the full range of variants indicated in different health conditions, or the variants that could drive research into alternative treatments or help understand the need for dose reductions.
Assuming white European genetics broadly apply to all people can ultimately lead to patients being mismanaged. Take the DPYD gene, as an example. This gene encodes for the enzyme dihydropyridine dehydrogenase (DPD), which is essential to the metabolism of the chemotherapy agents 5-fluoropyrimidine and capecitabine. The four variants of the DPYD gene currently tested for on the NHS, which would indicate that patients would have severe side effects and require dose modification, are based on data from trials conducted predominantly in people of white European ancestry[13]. In a case report, an Indian woman, aged 59 years, treated with full-dose capecitabine was admitted for severe mucositis of the ileum owing to a rare DPYD gene variant not previously identified[13]. Testing had only been carried out on the four main DPYD gene variants identified from the original research[14]. This poses the question: what if this gene variant is not as rare as we think and is simply undiscovered in people of Indian ancestry?
Given the impact that this lack of data can have on the patient care we offer and the direction of treatment innovation, it is vital that pharmacogenomic research represents diverse populations. To support researchers, frameworks should be provided which support community engagement — for example, how to improve community engagement and build patient and public trust[11]. These frameworks can be developed at a national level, involving patients and the public working alongside researchers, to support diversity in trial development.
Building trust
For people from different ancestral backgrounds to be represented within pharmacogenomic research, we must also overcome the hesitancy seen in some populations towards research[3]. Historical examples of racism within research cannot be ignored. The infamous Tuskegee Syphilis Project, which began in 1932, deliberately denied penicillin to African American men, a drug that became standard of care for syphilis in 1947[15]. The trial was only ended in 1972 after the details were leaked to the press and is just one of many examples of racism within research[15]. Building trust also requires absolute transparency and reassurance. As a healthcare community, we must ensure that a person’s DNA will be safely stored and used only for the purposes in which the consent was provided for. Lessons continue to be learnt through the story of Henrietta Lacks, an African American woman whose cervical cancer cells in 1951, now known as HeLa cells, were taken without her consent and continue to be used for medical research[16,17]. Stories like these continue to be passed down through generations, generating distrust in the healthcare profession and systems[3].
Another major barrier to consider is language; for example, can the words ‘pharmacogenomics’, ‘sequencing’ and ‘genotype’ be successfully translated into different languages? Apart from thinking of the technical elements of translating from one language to another, it is important to ensure cultural translation is undertaken, by having awareness of a community’s values, beliefs and customs[18].
There are some initiatives already underway that could be replicated elsewhere, such as the genomic cafes set up in Wales, which are informal spaces for people affected by genetic conditions to learn more about their health and discuss their needs[19]. In Bradford and Leeds, mental healthcare workshops and public health initiatives for childhood obesity prevention have been organised in mosques[20,21]. The same could be done with genomics, utilising places of worship or local community centres to help identify other barriers and build patient networks through the help of community leaders.
Population-wide testing
UK population demographics are shifting, with an increasing number of younger generations (18- to 24-year-olds) identifying themselves as belonging to two different ancestral groups, compared to people over the age of 60 years who self-identify within one group[22]. This supports the potential benefits of a population-based pharmacogenomic testing model. For example, HLA*15:02 is a gene associated with increased risk of life-threatening Steven-Johnson syndrome and toxic epidermal necrolysis (SJS/TENs) in patients treated with carbamazepine, and is commonly found in populations from Thailand, Taiwan, Singapore and China South Han[23]. In Thailand, HLA-B*15:02 testing has been introduced nationally as a preventive tool for SJS/TENs, with cases reducing from 1,300 (2019) to 300 (2021) per annum, and has led to new research initiatives within pharmacogenomics[24]. In the United States, broad population testing without ethnic pre-selection identified twice the number of HLA*15:02 carriers at risk of SJS/TENs[25]. This raises the question of whether broad testing is required in countries with greater diversity, and highlights that each country may need to adopt different pharmacogenomic screening models to reflect their population.
To reduce the risk of widening health inequalities, population-wide testing is a potential approach, which could simultaneously develop new research opportunities to fill data gaps, alongside population health economic models.
Looking ahead
Work has begun within Genomics England to ensure greater diversity within trial recruitment, with a diverse data initiative to increase representation across different ancestry groups to fill the data gaps within the UK genome database. However, there is still more action needed across genomics research by all those involved, including collaborative working with patients, carers and the public[25].
We all must improve our genomic literacy, to counsel all our patients on the merits and limitations of pharmacogenomics confidently
The establishment of seven Genomic Medicine Service Alliances (GMSAs) across all areas of England means that pharmacogenomics is now at the forefront of future service developments. These alliances will be critically appraising data and research to ensure diversity and inclusivity. For example, one GMSA priority within the DPYD national transformation project includes identifying relevant DPYD variants in patients of non-European ancestry; this will be achieved via ongoing research.
As pharmacy professionals, we also have some responsibility in this. We all must improve our genomic literacy, to counsel all our patients on both the merits and the limitations of pharmacogenomics confidently. Pharmacogenomics will not replace our clinical judgement; it is an additional tool within our medicines optimisation toolbox, but to use it effectively, testing and treatment must fully represent the populations we serve. Within pharmacy, our role will involve understanding the existing data gaps to identify where further research is needed to ensure equitable care and treatment within pharmacogenomics.
- 1Pharmacogenomics. Genomics Education Programme. https://www.genomicseducation.hee.nhs.uk/glossary/pharmacogenomics/ (accessed 7 Apr 2022).
- 2Fuentes A, Ackermann RR, Athreya S, et al. AAPA Statement on Race and Racism. Am J Phys Anthropol. 2019;169:400–2. doi:10.1002/ajpa.23882
- 3100,000 Genomes Project Black African and Black Caribbean Communities: A Qualitative Exploration of Views on Participation. INCLUSION BY DIALOGUE AND DESIGN. 2018.https://vdocument.in/inclusion-by-dialogue-and-design-the-100000-genomes-project-is-aiming-to-sequence.html?page=1 (accessed 7 Apr 2022).
- 4Ortega VE, Meyers DA. Pharmacogenetics: Implications of race and ethnicity on defining genetic profiles for personalized medicine. Journal of Allergy and Clinical Immunology. 2014;133:16–26. doi:10.1016/j.jaci.2013.10.040
- 5The anthropological study of education. Britannica. https://www.britannica.com/science/anthropology/The-anthropological-study-of-education (accessed 7 Apr 2022).
- 6Zhang H, De T, Zhong Y, et al. The Advantages and Challenges of Diversity in Pharmacogenomics: Can Minority Populations Bring Us Closer to Implementation? Clin. Pharmacol. Ther. 2019;106:338–49. doi:10.1002/cpt.1491
- 7Zhou Y, Ingelman-Sundberg M, Lauschke V. Worldwide Distribution of Cytochrome P450 Alleles: A Meta-analysis of Population-scale Sequencing Projects. Clin. Pharmacol. Ther. 2017;102:688–700. doi:10.1002/cpt.690
- 8McInnes G, Lavertu A, Sangkuhl K, et al. Pharmacogenetics at Scale: An Analysis of the UK Biobank. Clin. Pharmacol. Ther. 2020;109:1528–37. doi:10.1002/cpt.2122
- 9Magavern EF, Gurdasani D, Ng FL, et al. Health equality, race and pharmacogenomics. Brit J Clinical Pharma. 2021;88:27–33. doi:10.1111/bcp.14983
- 10Twesigomwe D, Wright GEB, Drögemöller BI, et al. A systematic comparison of pharmacogene star allele calling bioinformatics algorithms: a focus on CYP2D6 genotyping. npj Genom. Med. 2020;5. doi:10.1038/s41525-020-0135-2
- 11Fatumo S, Chikowore T, Choudhury A, et al. A roadmap to increase diversity in genomic studies. Nat Med. 2022;28:243–50. doi:10.1038/s41591-021-01672-4
- 12Population of England and Wales. UK Government. 2020.https://www.ethnicity-facts-figures.service.gov.uk/uk-population-by-ethnicity/national-and-regional-populations/population-of-england-and-wales/latest (accessed May 2022).
- 13Ly RC, Schmidt RE, Kiel PJ, et al. Severe Capecitabine Toxicity Associated With a Rare DPYD Variant Identified Through Whole-Genome Sequencing. JCO Precision Oncology. 2020;:632–8. doi:10.1200/po.20.00067
- 14Henricks LM, Lunenburg CATC, de Man FM, et al. DPYD genotype-guided dose individualisation of fluoropyrimidine therapy in patients with cancer: a prospective safety analysis. The Lancet Oncology. 2018;19:1459–67. doi:10.1016/s1470-2045(18)30686-7
- 15Saini A. Superior: The Return of Race Science. 1st ed. Fourth Estate 2019.
- 16Skloot R. The immortal life of Henrietta Lacks. Pan 2011.
- 17Henrietta Lacks: science must right a historical wrong. Nature. 2020;585:7–7. doi:10.1038/d41586-020-02494-z
- 18Schaafsma ES, Raynorr DK, de Jong‐van den Berg LTW. Pharmacy World and Science. 2003;25:185–90. doi:10.1023/a:1025812716177
- 19Genome UK: the future of healthcare. GOV.UK. 2020.https://www.gov.uk/government/publications/genome-uk-the-future-of-healthcare (accessed 7 Apr 2022).
- 20Dogra SA, Rai K, Barber S, et al. Delivering a childhood obesity prevention intervention using Islamic religious settings in the UK: What is most important to the stakeholders? Preventive Medicine Reports. 2021;22:101387. doi:10.1016/j.pmedr.2021.101387
- 21Mental Health Awareness Workshop at Leeds Grand Mosque. Touchstone. 2017.https://touchstonesupport.org.uk/mental-health-awareness-workshop-at-leeds-grand-mosque/ (accessed 7 Apr 2022).
- 22Ethnicity Facts and Figures: Age groups. GOV.UK. 2020.https://www.ethnicity-facts-figures.service.gov.uk/uk-population-by-ethnicity/demographics/age-groups/latest (accessed 7 Apr 2022).
- 23Fang H, Xu X, Kaur K, et al. A Screening Test for HLA-B∗15:02 in a Large United States Patient Cohort Identifies Broader Risk of Carbamazepine-Induced Adverse Events. Front. Pharmacol. 2019;10. doi:10.3389/fphar.2019.00149
- 24Bringing the Benefits of Genome Sequencing to the World. Public Policy Projects. https://publicpolicyprojects.com/wp-content/uploads/sites/6/2022/02/PPP-Global-Genomics-Report-122.pdf (accessed 7 Apr 2022).
- 25Diverse Data. Genomics England. https://www.genomicsengland.co.uk/initiatives/diverse-data (accessed 7 Apr 2022).