Abstract
Mental health conditions are the largest single cause of disability in the UK and among the top ten causes of disability worldwide. The prevalence of mental health conditions is increasing rapidly, especially since the COVID-19 pandemic. Overall, they cost the UK economy £118bn each year and are associated with a profound impact on people, their families and communities. Antidepressants and antipsychotics are the most common treatments for mental illnesses; however, these medications are associated with troublesome and often significant adverse drug reactions. It may be possible to identify those patients most at risk of developing severe side effects and those who are unlikely to respond to treatment using their genetics. Pharmacogenomics investigates the effects of inherited gene differences on the pharmacodynamics and pharmacokinetics of drugs and, subsequently, drug exposure and therapeutic effects. This article reviews the management of mental illnesses, and the role of genetics in disease risk and response to treatment. It will also discuss pharmacogenomic testing, guidelines and some of the barriers to its implementation in clinical settings.
Key words: Mental illness; antipsychotics; pharmacogenomics; pharmacogenomic testing; PGx
Introduction
The term ‘pharmacogenomics’ is often used interchangeably with ‘pharmacogenetics’ in association with precision medicine. The term ‘pharmacogenetics’ is an older term that has been in use since 1959. It refers to the idea that response to a drug is controlled by a person’s genes, particularly genes that determine drug metabolism[1,2]. As analytical methods improved, and the entire human genome was sequenced in 1997, the term ‘pharmacogenomics’ began to be used instead of pharmacogenetics[1]. It holds a similar meaning to pharmacogenetics, in that a person’s response to a drug is defined by their genetics; however, it explores the impact of a broader set of genes (not just metabolism genes) and non-coding regions of DNA on drug response[1,2].
Pharmacogenomics investigates the effects of inherited gene differences on the pharmacodynamics and pharmacokinetics of drugs and, subsequently, drug exposure and therapeutic effects, thereby improving the safety and therapeutic outcomes of pharmacological treatments[3–5]. This approach is vital to personalising treatment to reduce the likelihood or impact of severe adverse drug reactions, and to tailor drug choice and dosing to achieve the desired therapeutic outcomes[5]. Pharmacogenomics is a broad term that is associated with proteomics (i.e. the analysis of the whole protein set of a cell/tissue/ organelle) and metabolomics (i.e. the study of the entire set of metabolites within a cell/tissue/organelle for a specific cellular function)[6].
Mental health conditions are the largest single cause of disability in the UK and are among the top ten leading causes of disability worldwide[7,8]. In 2019, there were 10.3 million presentations of poor mental health in the UK and it is likely that 1 in 6 adults in England has experienced a common mental health condition, such as anxiety and depression, in the past week[9]. The COVID-19 pandemic has further increased the prevalence (and undoubtedly the impact) of mental health disorders. The prevalence of moderate to severe depressive symptoms rose from 10% at the start of the pandemic in March 2020 to 19% by March 2021[10]. Recent reports suggest that more people are now experiencing psychotic disorders, noting an increase from 0.4% in 2014 to 0.7% in 2017, and a 29% increase in referrals for first suspected episode of psychosis between 2019 and 2021[11,12]. Consequently, the prescribing of antipsychotics and antidepressants has increased significantly[13–15].
This article will review the management of mental health conditions and the role of genetics in disease risk and response to treatment, with a focus on antidepressants and antipsychotics. Pharmacogenomic testing, existing guidelines and some of the barriers to its implementation in clinical settings will also be discussed.
The burden of mental health conditions
Mental health conditions, especially severe mental illness (SMI), have a profound impact on people, their families, communities and the UK economy. On average, people with SMI die 15–20 years earlier than the general population; 2 in 3 of those deaths are from preventable physical illnesses, such as cardiometabolic and respiratory diseases[16]. Mental health conditions also cost the UK economy more than £117.9bn each year, which represents 5% of UK gross domestic product[9]. Most of these costs are from associated healthcare costs, loss of productivity (as people suffering from mental health conditions tend to work less or take time out of work) and the cost of support from informal carers[9].
The causes of mental illness are still subject to debate between those who suggest biological (e.g. altered neurotransmitter release and signalling), social (e.g. loneliness and stigma) and psychological (e.g. trauma) aetiologies are the driving factors, compared with those who consider mental illnesses as neurological disorders and those who see them as sociological conditions[17]. Many believe in a multi-factorial aetiology that is the result of a complex interaction between biology and the environment, including social and psychological factors — all of which require tailored interventions[17]. Understanding of the aetiology of mental illness is important, as it forms the basis of treatment and influences clinicians’ approaches to its management[18,19].
Is it all in the genes?
SMIs tend to cluster in families and can be hereditary[20]. It is estimated that heritability (i.e. the proportion of disease risk that can be attributed to genetic causes in a particular environment/ population) accounts for:
- 20–45% of anxiety disorders, obsessive-compulsive disorder, post-traumatic stress disorder and major depressive disorder;
- 50–60% of alcohol dependence and anorexia nervosa presentations;
- More than 75% of autism spectrum disorder, attention deficit hyperactivity disorder (ADHD), schizophrenia and bipolar disorders[20,21].
The genetic basis of most psychiatric disorders (e.g. ADHD, schizophrenia and bipolar disorder) is polygenic, which means that they are influenced by changes or variants in many genes. Each gene variant confers a small increase in risk but, cumulatively, the risk increases significantly[17].
Genetic variants implicated in mental illness include duplication or deletions of many nucleotide base[22]. The DNA sequence is altered when a single nucleotide is changed; for example, a nucleotide sequence of ‘ACG’ changes to ‘ACT’. If this single nucleotide change is found in at least 1% of the population, it is then referred to as a single nucleotide polymorphism (SNP). Notably, there is a growing shift towards using more accurate genomic terminologies, such as single nucleotide variant (SNV) instead of SNP, as SNVs can be rare in one population but common in another population, therefore making them more reflective of a particular variation[23]. However, for consistency with the cited literature, they will continue to be referred to as SNPs in this article.
A single alteration in nucleotide sequence may produce a different amino acid or cause an incomplete amino acid chain, resulting in a protein that cannot fulfil its normal function, with the potential to cause disease[24]. Individuals with a specific duplication of a portion of chromosome 16 (specifically in the 16p11.2 region) have been shown to have a 14-fold increased risk of psychosis and a 16-fold risk of developing schizophrenia[22]. Deletions on chromosomes 1, 15 and 22 are also associated with an increased risk of schizophrenia — these deletions collectively remain a rare cause of the condition[25]. Furthermore, SNPs affecting the dopamine reception D4 gene (DRD4), monoamine oxidase A gene (MAOA), 5-hydroxytryptamine receptor 1A gene (HTR1A) and other genes have also been associated with risks for major depressive disorder[26]. A comprehensive exploration of the SNPs implicated in SMIs and cognitive function is presented in a study by Golovina et al.[27].
Understanding mental illness through genome-wide association studies
Genome-wide association studies (GWAS) allow for comparison of the DNA sequence of populations with certain medical conditions against a healthy volunteer cohort, with the aim of identifying SNPs that may be associated with diseases[28]. If certain SNPs are more frequent in the group of participants with the disease, then these variations are suggested to be associated with the disease. Further studies are then carried out on the locus (i.e. location) of the variation to understand the genetic changes, as GWAS alone cannot establish causality between a genetic variant and disease[28].
More than 241 loci have been linked with SMIs, including schizophrenia, bipolar disorder, major depressive disorder and others[29]. Of the loci in question, 22 are implicated in at least 2 of the following illnesses: ADHD, alcohol dependence, anorexia nervosa, autism, bipolar disorder, major depression, obsessive-compulsive disorder, post-traumatic stress disorder, schizophrenia and Tourette’s syndrome. The ANK3, NDST3, and PLXNA2 loci have a strong association with both schizophrenia and bipolar disorders[30]. However, it is thought that schizophrenia and bipolar disorder result from a complex interplay of genetic polymorphisms and environmental factors (such as cannabis use, urbanisation and migration)[31]. The link between genetic polymorphism and the underlying biology of schizophrenia and bipolar disorder is complex and remains largely unknown[32].
Utilisation of GWAS also highlights the potential in targeting these genes/pathways for therapeutic management. Extensive efforts are underway to mine data available on drug–gene interactions to identify potential drug targets. The Drug–Gene Interaction Database mines a range of resources (such as DrugBank, PharmGKB, Chembl and Drug Target Commons) to help researchers annotate genes with known drug–gene interactions and develop hypotheses about how these genes form potential targets for drug development[33]. For example, Gaspar and Breen’s modelling study suggests that selective calcium channel blockers and antiseizure medicines (targeting GABA and glutamate receptors) may be good candidates for repurposing for use in schizophrenia management[34–36].
Antipsychotic and antidepressant medicines
Owing to their activity on a range of neurotransmitters, antipsychotic and antidepressant medicines are used to manage schizophrenia, bipolar disorder and many other SMIs. A recent review of the efficacy and tolerability of 32 antipsychotics found little differences between them in terms of their efficacy; however, their tendency to cause adverse drug reactions varies considerably from one agent to another[37].
Antipsychotics may be categorised as typical or atypical. Compared with atypical antipsychotics, typical antipsychotics have a stronger affinity to bind to dopaminergic receptors and ability, at a dose that is effective within the licensed dosage range, to induce extrapyramidal symptoms; for example, dystonia, pseudoparkinsonism (tremor and/or rigidity), akathisia (restlessness), and tardive dyskinesia (abnormal movements)[38]. Furthermore, atypical antipsychotics tend to have a higher affinity for serotonergic receptors[38]. Antipsychotics may also be called first-generation antipsychotics (FGAs) and second-generation antipsychotics (SGAs), based on the time of introduction into clinical practice, rather than on pharmacological activity or side effect profile[38].
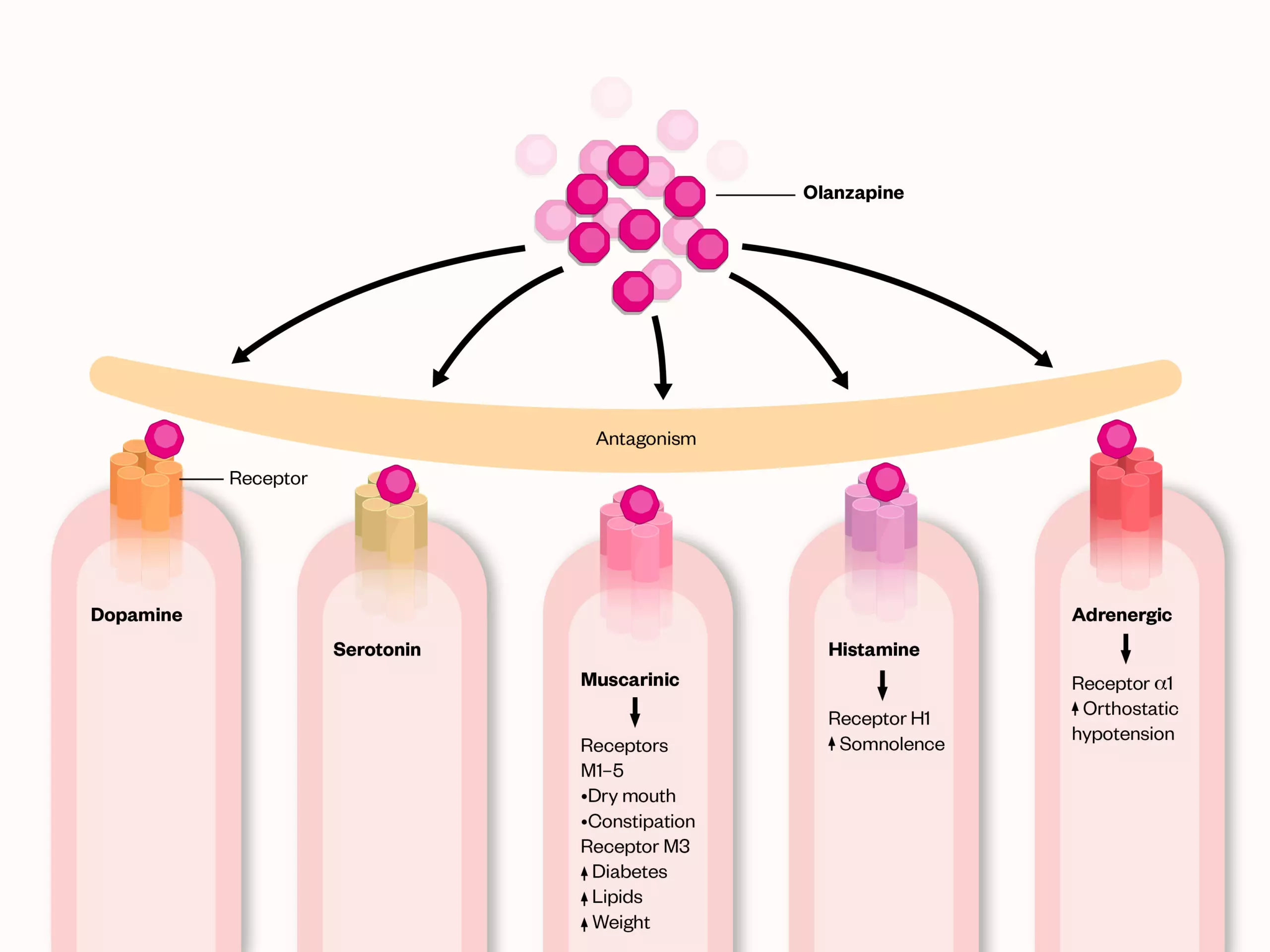
Antipsychotics act primarily through dopamine receptor antagonism, inhibiting neurotransmission at the dopamine receptors (D2 receptors) in the brain, which is particularly useful for the management of positive symptoms, referred to as ‘positive’ because they add on symptoms, such as hallucinations and delusions. They also block adrenergic, muscarinic and histamine receptors, causing a range of side effects[39]. Each antipsychotic has a unique set of side effects that can affect people differently[40]. However, FGAs (which usually include the typical antipsychotics) such as haloperidol and sulpiride are more likely to cause extrapyramidal motor side effects and hyperprolactinemia, whereas SGAs (atypical antipsychotics), such as amisulpride, paliperidone, and risperidone, are less likely to cause these side effects. SGAs, such as aripiprazole, are more likely to cause significant weight gain (cardiometabolic symptoms) and sedation[37].
Olanzapine is a SGA that is used in the management of schizophrenia, bipolar disorder and depression. Its mechanism of action (see Figure 1) represents the mode of action of most SGAs[41]. Patients are likely to be prescribed SGAs on a long-term basis, and their side effects will likely affect the patients’ cardiometabolic parameters, cognition, adherence and quality of life. Given that there is variability in efficacy (improving positive and negative symptoms) and side effects (mainly cardiometabolic and neurological, such as tardive dyskinesia) among individual antipsychotics, it is important to carefully consider the side effect profile of antipsychotic medicines to maintain a favourable risk–benefit ratio[37,38,42]. Antidepressants affect the availability/function of monoamine neurotransmitters (e.g. serotonin, noradrenaline and dopamine), by inhibiting the reuptake of these neurotransmitters (e.g. selective serotonin reuptake inhibitors ± noradrenaline and tricyclic antidepressants [TCAs]); exerting agonistic effects on post-synaptic receptors (e.g. vortioxetine) and blocking α2-adrenoceptors to enhance noradrenaline release (e.g. mirtazapine). Table 1 summarises the mode of action of many of the antidepressants on the market in the UK[43–45]. Similar to antipsychotics, antidepressants can also cause a range of adverse drug reactions, including prolongation of the QT interval, hypertension, bleeding (owing to inhibition of platelet aggregation), gastrointestinal side effects and dry mouth[46].
The limited efficacy and adverse effects of antidepressants and antipsychotics
Many patients may not respond to antipsychotic and antidepressant medication. It is thought that the prevalence of treatment resistance in psychiatric disorders ranges from 20–60%, owing to the varying definition of treatment resistance used across studies[47]. Reasons for treatment resistance are complex and often unclear. However, PGx testing-guided prescribing may improve treatment response and outcomes in those patients[48]. Treatment-resistant schizophrenia can occur in up to 34% of patients, whose symptoms persist despite a trial of two or more antipsychotic medicines at an adequate dose, for an adequate treatment duration and with confirmed adherence[49,50]. Various theories have been suggested to explain treatment resistance; one relates to the use of antipsychotics, and attributes resistance to the changes in dopaminergic pathways, as a result of exposure to dopamine antagonists. Blocking dopamine receptors (D2) for a prolonged period increases their density, which may cause the emergence of positive symptoms (e.g. psychosis). As a result, clinicians will often increase the antipsychotic medication to control the symptoms. Increasing the dose/medication will further increase the density of dopamine receptors, leading to increased dopamine supersensitivity and the re-emergence of positive symptoms[49]. It is also thought that treatment resistance is caused by changes in glutamate and serotonin transmission; however, the mechanisms involved are less clear[49].
Treatment resistance has also been reported in patients with major depressive disorder, where only 42–53% of patients respond to antidepressants[51]. This is thought to be owing to a complex interplay of factors, including P-glycoproteins in the blood–brain barrier, limiting the movement of drugs to the brain, and causing structural brain changes, altered levels of neurotransmitters (e.g. serotonin and glutamate) and subsequent changes in their signalling pathways[52,53]. Notably, research interest in treatment resistance is growing, but it remains scarce and hampered by the lack of consensus on definitions of treatment resistance, along with poor understanding of underlying mechanisms of resistance and the lack of effective interventions to manage it[47].
Pharmacokinetics and adverse drug reactions
Another factor that may limit the efficacy of antipsychotics and antidepressants relates to the pharmacokinetics of these medicines; the most relevant being their metabolism by the cytochrome P450 (CYP) isoenzymes. Most antipsychotics are metabolised by CYP1A2, CYP2C9, CYP2C19, CYP2D6 and CYP3A4 isoenzymes[54]. Variation in coding of the genes for these isoenzymes can result in altered enzyme activity (i.e. their ability to metabolise drugs), which can impact the level of drug exposure and their therapeutic effect[55].
Around 40% of antipsychotics involve the CYP2D6 isoenzyme in their metabolism, including aripiprazole, risperidone, and haloperidol[56,57]. Depending on the genetic variation of the CYP isoenzymes people carry, they can be:
- Poor metabolisers (PM);
- Intermediate metabolisers (IM);
- Normal metabolisers (NM);
- Ultra-rapid metabolisers (UM)[55].
Metaboliser status is CYP isoenzyme-specific. PMs and IMs tend to have higher drug levels in the body and are more likely to experience the drugs’ side effects, compared to NMs and UMs. The lower the ability to metabolise drugs, the higher the drug levels are in the body, increasing the risk of side effects/toxicity[58]. Note that this is relevant to active drugs as opposed to pro-drugs. A prodrug is a pharmacologically inactive drug that gets converted to an active drug through a chemical or enzymatic process[59].
Depending on the CYP2D6 genetic variant patients carry, side effects of antipsychotics include: treatment resistance (the term resistance here was used to suggest increased switching from risperidone to another antipsychotic when administrated to CYP2D6 PMs or UMs), hyperprolactinaemia, and increased length of hospital stay, with PMs and UMs staying longer in hospital[54,60]. The CYP2C19 genetic variants have been implicated in increased sedation with quetiapine and a higher risk of metabolic syndrome with clozapine[54].
The CYP2D6 and CYP2C19 enzymes are major metabolisers of most antidepressants. A meta-analysis of probability estimates by Koopmans et al. suggests that 36% of the population globally are likely to possess a CYP2D6 genetic variant, and that around 62% possess a CYP2C19 variant; these variants can lead to non-normal metabolism[61]. A study by Ricardo-Silgado et al. concluded that CYP2C19 PMs and IMs put on significantly more weight while taking citalopram compared to NMs and UMs[62]. Furthermore, CYP2B6 polymorphism (particularly CYP2B6*6) is suggested to affect the antidepressant and smoking cessation effects of bupropion, a noradrenaline and dopamine reuptake inhibitor[63].
The limited efficacy of antidepressants and antipsychotics in many patients, coupled with the burden of adverse drug reactions and the relapsing-remitting nature of mental illnesses, account for high rates of non-adherence to treatment, which have a subsequent impact on outcomes for these patients[64]. Pharmacogenomics has the potential to improve the safety and outcomes of antipsychotics and antidepressants through optimising drug choice and dosage to suit the patient’s variants[5,65]. Guidelines and prescribing support tools (such as the Canadian tool Sequence2Script) are being developed to integrate and/or streamline PGx testing, results and prescribing decisions to improve the therapeutic and safety outcomes of medicines[66]. In Canada, clinicians can enter the patient’s genetic information and their current medication into the Sequence2Script online tool, which will then generate a report highlighting suitable medication and recommended dosing based on the patient’s genetic information[67].
Approaches to pharmacogenomic testing and improved treatment outcomes
It is estimated that around 95% of the general population carry at least one actionable pharmacogenomic variant (i.e. a variant for which there is sufficient clinical evidence to support dose adjustment or alternative therapy)[68]. The chances of an individual being prescribed a drug affected by a pharmacogenomic variant are fairly high; a longitudinal study of primary care patients reported that around 60% of patients were prescribed more than two drugs affected by a pharmacogenomic variant over a 20-year period[69].
Pharmacogenomic variation can be tested for single or multiple genes, either reactively, by testing at the point of prescribing a specific drug, or pre-emptively, where results are held on the patient’s medical record to guide future prescribing decisions[70,71]. It is important to note that most approaches to pharmacogenomic testing interrogate the most common variants only, and the patient may still carry a rare variant with implications for drug metabolism, despite a ‘negative’ pharmacogenomic test result[72]. Furthermore, the pharmacogenomic results should always be considered in the context of a patient’s medical history and comorbidities[73]. Other factors, such as the effect of drug interactions and epigenetics, remain important considerations[74,75].
Pre-emptive screening, using a panel to detect variants across several genes for the most common pharmacogenomic variants, has the potential to reduce the risk of delay in commencing personalised treatment and may provide a cost-effective, efficient alternative to reactive testing of single gene variants each time a relevant drug is initiated[76–78]. However, the success of this approach relies on clinicians’ awareness of test results and the integration of healthcare records to allow healthcare professionals access to a patient’s results across all healthcare settings.
Variants in CYP2D6 and CYP2C19 in particular, can affect the metabolism of several antidepressants and pre-emptive screening for variants in multiple genes in patients with depression has shown promise[79,80]. The GUIDED study randomised 1,167 patients with major depressive disorder, who had inadequately responded to prior treatment, to receive either standard care or pharmacogenomic testing to guide medication selection[81]. For patients in the pharmacogenomic testing arm, genetic variants were screened across eight genes (CYP1A2, CYP2C9, CYP2C19, CYP3A4, CYP2B6, CYP2D6, HTR2A, SLC6A4). While pharmacogenomic testing did not improve mean depression symptom scores (primary outcome), a significant improvement was demonstrated in response and remission rates at eight weeks[81].
The recent PRIME clinical trial randomised 1,944 patients with major depressive disorder to receive either standard care or pre-emptive pharmacogenomic testing[82]. The study considered variation in CYP1A, CYP2B6, CYP2C19, CYP2C9, CYP3A4, CYP2D6, UGT1A4, SLC6A4 and HTR2A in relation to antidepressants. Pre-emptive testing was shown to reduce the prescription of medicines with predicted drug–gene interactions compared to standard care, but produced small and non-persistent benefits in symptom remission[82].
A 2021 Canadian health technology appraisal utilised systematic literature reviews of clinical and cost-effectiveness, coupled with patient and carer narratives to evaluate multi-gene testing and decision support tools in major depression[83]. A total of 14 studies were considered, totalling 3,497 patients, including 10 primary studies and 4 post-hoc analyses. The review concluded that effectiveness varied markedly between different gene panels, ranging from showing no difference in depression scores compared to standard treatment, to eliciting remission. The impact on adverse effects was determined to be uncertain. The overall evidence for multi-gene panel testing in major depression was determined as uncertain with low to very low confidence that the observed effects represented true effects[83]. The appraisal also reported that patients with major depression and their caregivers generally supported multi-gene pharmacogenomic testing, but some voiced concerns regarding confidentiality and of the possibility that pharmacogenomic guidance may replace patient-centred care[83]. As some trials to date have been underpowered or considered heterogenous interventions, it is hoped that larger prospective, randomised, controlled clinical trials will address these deficiencies, in addition to providing more comprehensive health economic data[84].
The Implementing Genomics in Practice network is undertaking a randomised trial of pre-emptive testing in patients with depression[85]. Patients will receive pre-emptive CYP2D6 and CYP2C19 testing, combined with clinical decision support, versus standard care followed by delayed pharmacogenomic testing at three months. The study aims to assess if genotype-guided antidepressant therapy results in improved control of depression assessed via patient-reported outcome tools. Given that patients remain at the centre of suffering from depressive illness, it is essential that their perception of the burden of depressive illness is assessed[86]. Effects on adverse effect burden and medication adherence will also be investigated[85].
Pharmacogenomic studies have investigated the link between genetic variants and clozapine-induced agranulocytosis (i.e. severely lowered white blood cell count). The ability to detect which patients are at lower risk of developing clozapine-induced agranulocytosis, by the absence of specific genetic variants, may allow a reduction in haematological monitoring[87]. However, the complex aetiology of clozapine-induced agranulocytosis and likely implication of variants of multiple genes, including HLA-DQB1, HLA-B and SLCO1B3/SLCO1B7, have created challenges in producing a pharmacogenomic test with appropriate sensitivity, specificity and adequate negative and positive predictive value[88]. A recent systematic review and meta-analysis also found that individuals with the HLA-DRB1*04:02 genetic variant had a nearly six-fold increase of developing clozapine-induced agranulocytosis[87]. Although authors note the relatively poor positive predictive value (versus a strong negative predictive value), the two studies of the HLA-DRB1*04:02 variant included in the meta-analysis comprised relatively low patient numbers and considered only non-Jewish German and Ashkenazi Jewish populations. The authors emphasise that further work is necessary to determine the clinical utility of testing in wider ancestral populations, given that the frequency and predictive value for agranulocytosis for the HLA-DRB1*04:02 variant may differ in patients of other ancestries[87]. A role for genomic testing for benign ethnic neutropenia has also been evaluated[89]. This would identify patients with neutropenia unrelated to clozapine, for whom monitoring parameters may be amended with the aim of reducing treatment interruptions[89].
A possible link between pharmacogenomic variants and an increased risk of tardive dyskinesia in patients prescribed antipsychotics has also been the subject of substantial research, with conflicting conclusions that, again, may not be applicable across different ancestries[90–94]. A meta-analysis of patients categorised as CYP2D6 poor metabolisers, demonstrated an increased risk of tardive dyskinesia that failed to reach statistical significance[90]. While a further meta-analysis found no significant association overall with CYP2D6 or CYP1A2 variants in either Asian or European populations, when analysis was confined to prospective studies only, a statistically significant link was found for CYP2D6[94]. A study in 516 American patients with SMIs highlighted links between variants in the DRD3 gene, which codes for a subtype of dopamine receptor and severe tardive dyskinesia, and the deletion of the GSTM1 gene and tardive dyskinesia[93]. A recent large GWAS of more than 1,400 participants discovered possible associations for variants in TNFRSF1B and CALCOCO1 — authors advise further investigation is warranted[95]. As TNFRSF1B is a member of the tumour necrosis factor receptor superfamily, this may implicate inflammatory processes in the pathogenesis of tardive dyskinesia and the authors also note that CALCOCO1 may play a role in regulating the activity of CYP P450 isoenzymes[95].
Pre-publication results of the ‘Pre-emptive Pharmacogenomic Testing for Preventing Adverse Drug Reactions’ (PREPARE) clinical trial demonstrate an impressive benefit for pre-emptive pharmacogenomic testing in reducing adverse drug reactions across a range of prescribed medicines, including antidepressants and antipsychotics[71,96]. This is the first randomised, prospective, international multi-centre trial of pre-emptive testing in Europe. It randomised nearly 7,000 patients to standard of care or pre-emptive testing, followed by dose adjustment or alternative treatment. Variants across 13 genes relating to more than 40 drugs, including selective serotonin reuptake inhibitors (SSRIs), TCAs and antipsychotics were considered. Pre-publication results indicate that adverse drug reactions were reduced by 30% by pre-emptive pharmacogenomic testing[96].
Pharmacogenomic dosing guidelines for antidepressants and antipsychotics
For the successful implementation of pharmacogenomics in mental health management, clear clinical guidelines are necessary to translate genomic laboratory test results into actionable prescribing advice.
Pharmacogenomic dosing guidelines have been produced by a range of European and international expert groups, including the Dutch Pharmacogenetics Working Group (DPWG), the French National Network (Réseau) of Pharmacogenetics (RNPGx), the Canadian Pharmacogenomics Network for Drug Safety (CPNDS) and the Clinical Pharmacogenetics Implementation Consortium (CPIC)[97–101]. The Pharmacogenomics Knowledgebase (PharmGKB) summarises and grades evidence for drug–gene pairs and signposts to dosing guidance and drug label annotations[102].
CPIC is an international group, mainly consisting of volunteers, which aims to facilitate pharmacogenomics use[100]. CPIC creates and curates peer-reviewed, evidence-based clinical practice guidelines for drug–gene combinations, focusing on providing information where genotype is known as opposed to advising if or when genotyping should be undertaken. The guidelines translate genotypes detailing the presence or absence of relevant genomic variants, into phenotypes or metaboliser status, for which dosing guidance is then provided. This is demonstrated in Figure 2, using citalopram as an example of an antidepressant potentially affected by variants of CYP2C19.
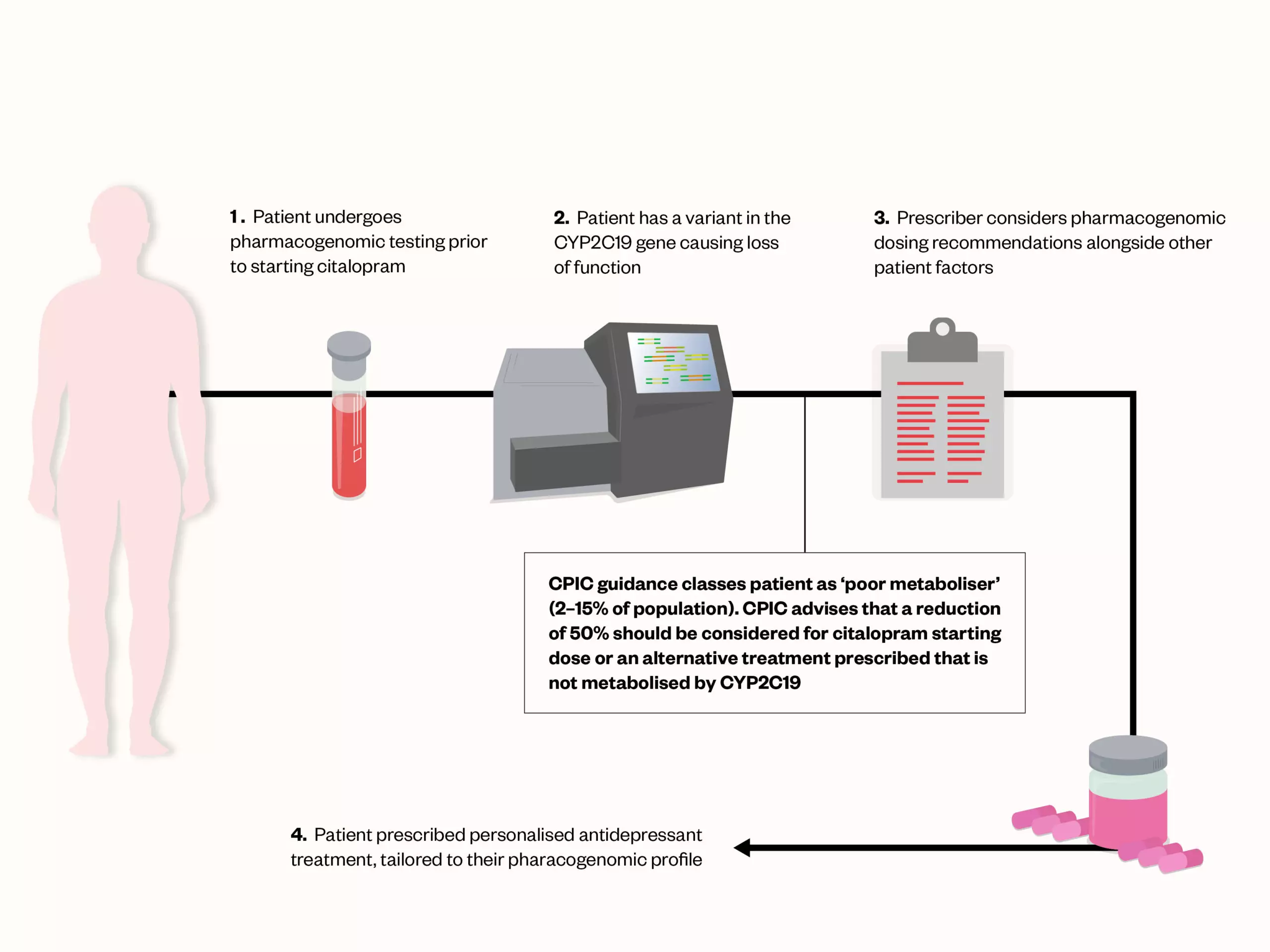
prescribed citalopram
There are currently CPIC pharmacogenomic guidelines supported by level A evidence for TCAs and SSRIs, detailed in Table 2[79,80]. Further CPIC guidelines are either planned or in development for antipsychotics and further classes of antidepressants. In areas where evidence is weaker, guidance may be conflicting, and the phenotype or metaboliser status may be updated as new evidence emerges. In addition to clinical guidelines, pharmacogenomic annotations are being incorporated into licensing information by several international bodies, including the US Food and Drug Administration, European Medicines Agency and Health Canada, which may support clinical implementation[103–105].
Pharmacogenomics and the NHS
Currently, within the NHS, widespread pharmacogenomic testing is not embedded into routine practice, except for DPYD testing for the gene encoding the dihydropyrimidine dehydrogenase enzyme prior to fluoropyrimidine chemotherapy and HLA-B screening for abacavir[106]. It is envisaged that PGx testing will become part of mainstream care in the future, with a recent report on personalised prescribing from the Royal College of Physicians and the British Pharmacological Society joint working party, acknowledging that this is likely to be an iterative process[70,107,108].
Both TCAs and SSRIs are included in the PROGRESS project, a pharmacogenomics pilot project in primary care supported by the NHS Genomic Medicine Service[109]. The PROGRESS project aims to assess the feasibility of embedding pharmacogenomic testing in primary care and to generate health economic data for use within the NHS setting. As outlined in the recent NHS England Genomics strategy, data from this, and other pilot projects, will inform wider pharmacogenomics implementation in England[70].
Barriers to the widespread use of pharmacogenomics within mental health
There are no international examples of the implementation of PGx testing in countries with publicly- funded healthcare systems and evidence of cost-effectiveness is lacking[107]. However, by reviewing the challenges faced in other countries, potential barriers and challenges to implementation in mental health can be surmised.
A recent review by Jameson et al. identified several themes as potential barriers to implementing PGx in mental health[64]. These include financial costs, lack of knowledge about PGx and the likely challenging integration of PGx into existing workflows, which is further explained in the following section.
Cost-effectiveness of pharmacogenomic testing in mental health
A systematic review of various cost-effectiveness studies highlighted that the most common mental health conditions in economic evaluation studies of PGx were schizophrenia and major depressive disorder[110]. Overall, although the cost-effectiveness of PGx testing appeared positive, these studies varied in design and effectiveness measures, making like-for-like comparisons challenging[110]. Furthermore, a qualitative study in primary care identified concerns regarding the cost-effectiveness of PGx testing, and the need to make informed commissioning decisions based on cost-effectiveness (i.e. whether PGx is good value for money), financial costs (i.e. extra costs associated with a new venture), and opportunity costs (missing out on the benefits of choosing one option over another)[111]. Results from a meta-analysis and systematic review of PGx-guided therapy in depressive symptom remission indicated that PGx testing provided a modest yet significantly favourable effect on improving symptoms[65]. When considering the loss of productivity associated with poor symptom control, along with the increased utilisation of healthcare resources, and the costs of managing adverse drug reactions, there are likely to be cost savings across the board[112].
A more recent systematic review of the cost-effectiveness of PGx testing with CPIC guidance indicated that 71% of studies evaluated were determined to be cost-effective or cost-saving[113]. It is important to acknowledge that most of the literature available for PGx testing in mental health is largely based on information from the United States. One of the main differences, apart from treatment options and guidelines in some cases, between the UK and the United States, is that the UK has publicly-funded healthcare systems, thus direct transferability of health economic findings may not be applicable.
Ongoing or recently completed research studies will assess the clinical outcomes, both in terms of treatment efficacy and adverse effects, experienced by patients who received PGx-guided mental health prescribing compared to standard care[70,114]. This forthcoming data; for example, from the PREPARE trial, which includes pharmacogenomic testing for antidepressants, antipsychotics and psychostimulants, will be valuable in determining the implementation strategy of PGx testing in psychiatry within the NHS in the UK[115].
Workforce and workflow
Once PGx testing is integrated into workflows and undertaken in the UK, the sample will need to be processed by NHS-approved laboratories, followed by results analysis and interpretation into a therapeutically meaningful recommendation. These data would need to be stored securely and relayed to the healthcare professional for further action. The results/recommendations should be underpinned by dosing guidelines and accessible to clinicians, who can make informed prescribing decisions while also considering other patient and clinical factors. For mental health conditions, informed consent could be complex owing to the patients’ symptoms. Furthermore, the PGx test results could open the door for further ethical considerations, especially if the findings are relevant to family members[116]. It is important to note that PGx findings for mental health conditions are highly likely to be relevant for other conditions/disciplines as well; for example, the CYP2D6 gene codes for the enzyme that metabolises medicines other than those used for mental health conditions, such as diabetes, cancer and cardiovascular medicines. Therefore, PGx will need to be implemented in a “cross-pathway” manner to avoid duplication of efforts and costs and enhance the utility of findings[117].
For this cross-pathway approach to work, pharmacogenomic test results should be accessible across different care interfaces, and especially so within mental health. Typically, depending on the condition and treatment required, prescribing may sometimes be initiated by a consultant psychiatrist in secondary care and then transferred to a primary care prescriber via a shared-care pathway. Therefore, the patient’s information needs to be visible systemwide, to a range of healthcare professionals in both primary and secondary care, who are adequately trained to interpret results against dosing guidance and relay results to the patient. The added challenge in mental health is that treatment usually involves multiple healthcare professionals, across a variety of settings, usually each using their own IT system. Furthermore, mental health bed shortages can result in patients being sent hundreds of miles away for treatment (out of area placement) and they may be placed in private hospitals, exacerbating the issue of accessible healthcare records[118]. Currently, electronic healthcare systems are not adequately integrated, and laboratory capacity, bioinformatic analysis and the education and training of the NHS workforce still requires substantial improvement[64].
Validated pharmacogenomic gene panels, diversity and patient concerns
Different populations exhibit differences in pharmacokinetics and pharmacodynamics and specific drug–gene pairs still need to be validated for use in mental health conditions; this data should be representative of diverse populations. Commercially available and direct to consumer (DTC) testing kits, which may offer pharmacogenomic testing to drug metabolism (e.g. antidepressants), or pre-disposition to mental health conditions (e.g. anxiety), may not be explicit in which variants are tested for and may not be applicable or appropriate for different population groups[66]. The DTC test panels may include genes where there is insufficient evidence to guide psychiatric prescribing e.g. DRD2 and CYP1A2[119]. Additionally, in mental health prescribing, the most relevant genes, as discussed earlier, include CYP2D6, CYP2C19 and CYP2C9; however, even when the same genes appear on a testing panel, there could be substantial variation in the number of sequence variations, or alleles, assayed within those genes[120]. Thus, where possible, standardised evidence-based panels should be utilised instead[121].
Genomic studies have predominantly enrolled patients of European descent, and this could influence clinical interpretation[122,123]. Within the UK Biobank 500,000 Genome Project, approximately 16% of data is from “non-white British” ancestry[124]. Efforts should be made to enhance recruitment of diverse participants to provide information that is relevant to the diverse populations treated in clinical settings.
Many populations are under-represented in clinical and genetic studies; therefore, patients from these under-represented populations may worry that the treatment options offered may not demonstrate the same effectiveness as in the studied population/genotype[125–127]. It is possible that, in the future, new treatments will be reserved for those with a particular genotype, in which a drug demonstrates better effectiveness[64].
Other patient concerns include how data could be used in the future and whether it could lead to employment, medical treatment or health insurance discrimination, owing to potential preclusion on disclosure of predisposing genetic conditions; storage of confidential information; and the use of technical medical language that would prevent patients from understanding PGx[128,129]. Although there are several resources available for healthcare professionals, patient-facing resources that adequately address patients’ concerns remain lacking.
While it is important to note concerns, there is also widespread optimism and enthusiasm in the power of PGx testing and its role in personalised medicine. There is acceptance, among patients and clinicians, of PGx’s ability to reduce adverse drug reactions, and improve precision prescribing and patients’ outcomes. There is also hope that PGx testing will become part of routine practice, in both primary and secondary care in the future[64,111,130–133].
Lack of UK PGx evidence-based prescribing guidance
Whilst UK national dosing guidance is available for DPYD pharmacogenomic testing (oncology), this is lacking in psychiatry[134]. Currently, UK mental health guidelines do not include prescribing or treatment recommendations based on PGx testing, thus, there is a lack of validated, pharmacogenomic-guided, treatment algorithms in psychiatry[130–133]. However, a 2021 guideline on the use of PGx for the selection of appropriate antidepressant or antipsychotic dose according to metaboliser status could be considered for adoption in UK practice. This a clinical guideline produced for prescribing psychotropic medicines, based on current information from DPWG and CPIC, which summarises the relevant literature and provides general recommendations for psychotropic prescribing and specific recommendations for specific psychotropic drug class, including CYP2C19 and CYP2D6 — the two most common genes in drug–gene pairing in psychiatry[55].
Looking ahead
The future of pharmacogenomics’ implementation in the UK is promising. In October 2022, the UK launched the first ever NHS genomics strategy ‘Accelerating genomic medicine in the NHS’. The strategy envisages developing a pioneering service delivery model, over the next five years, where genomics drives the prediction, prevention, diagnosis and management of diseases through precision medicine[70]. Early in 2023, a pilot programme will take place in the north west of England, where five general practices will be pharmacogenomic testing patients who are about to start a new statin, certain antidepressants or proton pump inhibitors, or have a change of medicine, to ensure they are prescribed the correct treatment option and reduce the risk of adverse drug reactions[109]. If successful, the service may become available nationally. The potential benefits of such offering to improving patient safety and outcomes could not be overstated in the ‘Personalised prescribing: using pharmacogenomics to improve patient outcomes’ report by the Royal College of Physicians and British Pharmacological Society[107]. Despite the optimism, the implementation of PGx testing within the NHS will require a multidisciplinary approach and potentially span across multiple specialities — normalising this offering will likely be challenging[107]. Crucially, patients’ health care records must be integrated between the primary and secondary interface, where pharmacogenomic information, alongside other information relevant to prescribing, is accessible to clinicians across the care pathway through summary care records.
The Royal Pharmaceutical Society highlights the potential role of pharmacists within pharmacogenomics, and its benefits to patients and multidisciplinary teams working within the NHS[108]. One of the fundamentals of implementation, apart from the physical testing infrastructure, is building capability and capacity within the pharmacy workforce. This can be achieved through education and training of mental health pharmacists in genomic medicine to enhance their input into clinical research, the development of infrastructure, health economics and governance of PGx testing within mental health services.
Conclusions
The prevalence of mental illnesses is on the rise, and the costs associated with their care are straining health budgets. The management of these conditions is long and complex and, if not optimised, can exacerbate the burden of symptoms and adverse reactions associated with treatment. Pharmacogenomics offers the potential to identify risk factors associated with sub-optimal responses to treatment, and even prevent severe and debilitating adverse drug reactions. Although the implementation of pharmacogenomics for mental health is still in its infancy, there is potential for significant benefits to patients in the future.
- 1Daly AK. Pharmacogenetics: a general review on progress to date. British Medical Bulletin. 2017;:1–15. doi:10.1093/bmb/ldx035
- 2Pirmohamed M. Pharmacogenetics and pharmacogenomics. British Journal of Clinical Pharmacology. 2001;52:345–7. doi:10.1046/j.0306-5251.2001.01498.x
- 3Farinde A. Overview of Pharmacodynamics. MSD Manual Professional Version. 2022.https://www.msdmanuals.com/en-gb/professional/clinical-pharmacology/pharmacodynamics/overview-of-pharmacodynamics (accessed Jan 2023).
- 4Le J. Overview of Pharmacokinetics. MSD Manual Professional Version. 2022.https://www.msdmanuals.com/en-gb/professional/clinical-pharmacology/pharmacokinetics/overview-of-pharmacokinetics (accessed Jan 2023).
- 5Rollinson V, Turner R, Pirmohamed M. Pharmacogenomics for Primary Care: An Overview. Genes. 2020;11:1337. doi:10.3390/genes11111337
- 6Walton TEF, Rajan JN. Omics and anaesthesia: pharmacogenomics, proteomics and metabolomics. Anaesthesia & Intensive Care Medicine. 2019;20:248–53. doi:10.1016/j.mpaic.2019.01.019
- 7NICEimpact mental health. National Institute for Health and Care Excellence. 2019.https://www.issup.net/files/2019-05/NICEimpact-mental-health.pdf (accessed Jan 2023).
- 8Global, regional, and national burden of 12 mental disorders in 204 countries and territories, 1990–2019: a systematic analysis for the Global Burden of Disease Study 2019. The Lancet Psychiatry. 2022;9:137–50. doi:10.1016/s2215-0366(21)00395-3
- 9McDaid D, Park A-L, Wahlbeck K. The Economic Case for the Prevention of Mental Illness. Annu. Rev. Public Health. 2019;40:373–89. doi:10.1146/annurev-publhealth-040617-013629
- 10Coronavirus and depression in adults, Great Britain: July to August 2021. Office for National Statistics. 2021.https://www.ons.gov.uk/peoplepopulationandcommunity/wellbeing/articles/coronavirusanddepressioninadultsgreatbritain/julytoaugust2021 (accessed Jan 2023).
- 11Mental health statistics (England). House of Commons Library. 2021.https://researchbriefings.files.parliament.uk/documents/SN06988/SN06988.pdf (accessed Jan 2023).
- 12Symptoms of psychosis significantly increased during pandemic. Rethink Mental Illness. 2021.https://www.rethink.org/news-and-stories/news/2021/10/symptoms-of-psychosis-significantly-increased-during-pandemic/ (accessed Jan 2023).
- 13Shoham N, Cooper C, Lewis G, et al. Temporal trends in psychotic symptoms: Repeated cross-sectional surveys of the population in England 2000–14. Schizophrenia Research. 2021;228:97–102. doi:10.1016/j.schres.2020.11.057
- 14Iacobucci G. NHS prescribed record number of antidepressants last year. BMJ. 2019;:l1508. doi:10.1136/bmj.l1508
- 15Rabeea SA, Merchant HA, Khan MU, et al. Surging trends in prescriptions and costs of antidepressants in England amid COVID-19. DARU J Pharm Sci. 2021;29:217–21. doi:10.1007/s40199-021-00390-z
- 16Severe mental illness (SMI) and physical health inequalities: briefing. Public Health England. 2018.https://www.gov.uk/government/publications/severe-mental-illness-smi-physical-health-inequalities/severe-mental-illness-and-physical-health-inequalities-briefing (accessed Jan 2023).
- 17Bhugra D, Bhui K, Yeung Shan Wong S, et al., editors. Oxford Textbook of Public Mental Health. Oxford University Press 2018. doi:10.1093/med/9780198792994.001.0001
- 18White F. Application of Disease Etiology and Natural History to Prevention in Primary Health Care: A Discourse. Med Princ Pract. 2020;29:501–13. doi:10.1159/000508718
- 19Kimmerle J, Anikin A, Bientzle M. The Impact of Perceived Etiology, Treatment Type, and Wording of Treatment Information on the Assessment of Gastritis Treatments. Front. Public Health. 2020;8. doi:10.3389/fpubh.2020.00035
- 20Smoller JW, Andreassen OA, Edenberg HJ, et al. Psychiatric genetics and the structure of psychopathology. Mol Psychiatry. 2018;24:409–20. doi:10.1038/s41380-017-0010-4
- 21Sullivan PF, Daly MJ, O’Donovan M. Genetic architectures of psychiatric disorders: the emerging picture and its implications. Nat Rev Genet. 2012;13:537–51. doi:10.1038/nrg3240
- 22Giaroli G, Bass N, Strydom A, et al. Does rare matter? Copy number variants at 16p11.2 and the risk of psychosis: A systematic review of literature and meta-analysis. Schizophrenia Research. 2014;159:340–6. doi:10.1016/j.schres.2014.09.025
- 23Sequence Variant Nomenclature. HGVS.org. 2022.https://varnomen.hgvs.org/bg-material/glossary/ (accessed Jan 2023).
- 24SINGLE NUCLEOTIDE POLYMORPHISMS (SNPS). National Human Genome Research Institute. 2023.https://www.genome.gov/genetics-glossary/Single-Nucleotide-Polymorphisms (accessed Jan 2023).
- 25Kirov G. The role of copy number variation in schizophrenia. Expert Review of Neurotherapeutics. 2010;10:25–32. doi:10.1586/ern.09.133
- 26Shadrina M, Bondarenko EA, Slominsky PA. Genetics Factors in Major Depression Disease. Front. Psychiatry. 2018;9. doi:10.3389/fpsyt.2018.00334
- 27Golovina E, Vickers MH, Erb CD, et al. GWAS SNPs Impact Shared Regulatory Pathways Amongst Multimorbid Psychiatric Disorders and Cognitive Functioning. Front. Psychiatry. 2020;11. doi:10.3389/fpsyt.2020.560751
- 28Genome-Wide Association Studies Fact Sheet. National Human Genome Research Institute . 2022.https://www.genome.gov/about-genomics/fact-sheets/Genome-Wide-Association-Studies-Fact-Sheet (accessed Jan 2023).
- 29Sullivan PF, Geschwind DH. Defining the Genetic, Genomic, Cellular, and Diagnostic Architectures of Psychiatric Disorders. Cell. 2019;177:162–83. doi:10.1016/j.cell.2019.01.015
- 30Prata DP, Costa-Neves B, Cosme G, et al. Unravelling the genetic basis of schizophrenia and bipolar disorder with GWAS: A systematic review. Journal of Psychiatric Research. 2019;114:178–207. doi:10.1016/j.jpsychires.2019.04.007
- 31Robinson N, Bergen SE. Environmental Risk Factors for Schizophrenia and Bipolar Disorder and Their Relationship to Genetic Risk: Current Knowledge and Future Directions. Front. Genet. 2021;12. doi:10.3389/fgene.2021.686666
- 32Mould AW, Hall NA, Milosevic I, et al. Targeting synaptic plasticity in schizophrenia: insights from genomic studies. Trends in Molecular Medicine. 2021;27:1022–32. doi:10.1016/j.molmed.2021.07.014
- 33Freshour SL, Kiwala S, Cotto KC, et al. Integration of the Drug–Gene Interaction Database (DGIdb 4.0) with open crowdsource efforts. Nucleic Acids Research. 2020;49:D1144–51. doi:10.1093/nar/gkaa1084
- 34Berger SM, Bartsch D. The role of L-type voltage-gated calcium channels Cav1.2 and Cav1.3 in normal and pathological brain function. Cell Tissue Res. 2014;357:463–76. doi:10.1007/s00441-014-1936-3
- 35Gaspar HA, Breen G. Drug enrichment and discovery from schizophrenia genome-wide association results: an analysis and visualisation approach. Sci Rep. 2017;7. doi:10.1038/s41598-017-12325-3
- 36Lago SG, Bahn S. The druggable schizophrenia genome: from repurposing opportunities to unexplored drug targets. npj Genom. Med. 2022;7. doi:10.1038/s41525-022-00290-4
- 37Huhn M, Nikolakopoulou A, Schneider-Thoma J, et al. Comparative efficacy and tolerability of 32 oral antipsychotics for the acute treatment of adults with multi-episode schizophrenia: a systematic review and network meta-analysis. The Lancet. 2019;394:939–51. doi:10.1016/s0140-6736(19)31135-3
- 38Schizophrenia and Related Psychoses. The Maudsley Prescribing Guidelines in Psychiatry. 2021;:1–224. doi:10.1002/9781119870203.mpg001
- 39Chokhawala K, Stevens L. Antipsychotic Medications. National Library of Medicine. 2022.https://www.ncbi.nlm.nih.gov/books/NBK519503/ (accessed Jan 2023).
- 40Stroup TS, Gray N. Management of common adverse effects of antipsychotic medications. World Psychiatry. 2018;17:341–56. doi:10.1002/wps.20567
- 41Boyle KR, Boyle JG, Fisher M, et al. Olanzapine. Pract Diab. 2014;31:304–305a. doi:10.1002/pdi.1891
- 42Velligan DI, Sajatovic M, Hatch A, et al. Why do psychiatric patients stop antipsychotic medication? A systematic review of reasons for nonadherence to medication in patients with serious mental illness. PPA. 2017;Volume 11:449–68. doi:10.2147/ppa.s124658
- 43Schulz P, Macher J-P. The clinical pharmacology of depressive states. Dialogues in Clinical Neuroscience. 2002;4:47–56. doi:10.31887/dcns.2002.4.1/pschulz
- 44Sheffler Z, Patel P, Abdijadid S. Antidepressants. National Library of Medicine. 2022.https://www.ncbi.nlm.nih.gov/books/NBK538182/ (accessed Jan 2023).
- 45British National Formulary. National Institute for Health and Care Excellence. 2022.https://bnf.nice.org.uk/ (accessed Jan 2023).
- 46Wang S-M, Han C, Bahk W-M, et al. Addressing the Side Effects of Contemporary Antidepressant Drugs: A Comprehensive Review. Chonnam Med J. 2018;54:101. doi:10.4068/cmj.2018.54.2.101
- 47Howes OD, Thase ME, Pillinger T. Treatment resistance in psychiatry: state of the art and new directions. Mol Psychiatry. 2021;27:58–72. doi:10.1038/s41380-021-01200-3
- 48Bousman CA, Arandjelovic K, Mancuso SG, et al. Pharmacogenetic tests and depressive symptom remission: a meta-analysis of randomized controlled trials. Pharmacogenomics. 2019;20:37–47. doi:10.2217/pgs-2018-0142
- 49Potkin SG, Kane JM, Correll CU, et al. The neurobiology of treatment-resistant schizophrenia: paths to antipsychotic resistance and a roadmap for future research. npj Schizophr. 2020;6. doi:10.1038/s41537-019-0090-z
- 50Siskind D, Orr S, Sinha S, et al. Rates of treatment-resistant schizophrenia from first-episode cohorts: systematic review and meta-analysis. Br J Psychiatry. 2021;220:115–20. doi:10.1192/bjp.2021.61
- 51Taliaz D, Spinrad A, Barzilay R, et al. Optimizing prediction of response to antidepressant medications using machine learning and integrated genetic, clinical, and demographic data. Transl Psychiatry. 2021;11. doi:10.1038/s41398-021-01488-3
- 52El-Hage W, Leman S, Camus V, et al. Mechanisms of antidepressant resistance. Front. Pharmacol. 2013;4. doi:10.3389/fphar.2013.00146
- 53Caraci F, Calabrese F, Molteni R, et al. International Union of Basic and Clinical Pharmacology CIV: The Neurobiology of Treatment-resistant Depression: From Antidepressant Classifications to Novel Pharmacological Targets. Pharmacol Rev. 2018;70:475–504. doi:10.1124/pr.117.014977
- 54Arranz MJ, Salazar J, Hernández MH. Pharmacogenetics of antipsychotics: Clinical utility and implementation. Behavioural Brain Research. 2021;401:113058. doi:10.1016/j.bbr.2020.113058
- 55van Westrhenen R, van Schaik RHN, van Gelder T, et al. Policy and Practice Review: A First Guideline on the Use of Pharmacogenetics in Clinical Psychiatric Practice. Front. Pharmacol. 2021;12. doi:10.3389/fphar.2021.640032
- 56Fleeman N, Dundar Y, Dickson R, et al. Cytochrome P450 testing for prescribing antipsychotics in adults with schizophrenia: systematic review and meta-analyses. Pharmacogenomics J. 2010;11:1–14. doi:10.1038/tpj.2010.73
- 57Cacabelos R, Cacabelos P, Aliev G. Genomics of schizophrenia and pharmacogenomics of antipsychotic drugs. OJPsych. 2013;03:46–139. doi:10.4236/ojpsych.2013.31008
- 58Wannasuphoprasit Y, Andersen SE, Arranz MJ, et al. CYP2D6 Genetic Variation and Antipsychotic-Induced Weight Gain: A Systematic Review and Meta-Analysis. Front. Psychol. 2022;12. doi:10.3389/fpsyg.2021.768748
- 59Cho S, Yoon Y-R. Understanding the pharmacokinetics of prodrug and metabolite. Transl Clin Pharmacol. 2018;26:1. doi:10.12793/tcp.2018.26.1.1
- 60Nofziger C, Turner AJ, Sangkuhl K, et al. PharmVar GeneFocus: CYP2D6. Clin. Pharmacol. Ther. 2019;107:154–70. doi:10.1002/cpt.1643
- 61Koopmans AB, Braakman MH, Vinkers DJ, et al. Meta-analysis of probability estimates of worldwide variation of CYP2D6 and CYP2C19. Transl Psychiatry. 2021;11. doi:10.1038/s41398-020-01129-1
- 62Ricardo-Silgado ML, Singh S, Cifuentes L, et al. Association between CYP metabolizer phenotypes and selective serotonin reuptake inhibitors induced weight gain: a retrospective cohort study. BMC Med. 2022;20. doi:10.1186/s12916-022-02433-x
- 63Wang P-F, Neiner A, Kharasch ED. Stereoselective Bupropion Hydroxylation by Cytochrome P450 CYP2B6 and Cytochrome P450 Oxidoreductase Genetic Variants. Drug Metab Dispos. 2020;48:438–45. doi:10.1124/dmd.119.090407
- 64Jameson A, Fylan B, Bristow GC, et al. What Are the Barriers and Enablers to the Implementation of Pharmacogenetic Testing in Mental Health Care Settings? Front. Genet. 2021;12. doi:10.3389/fgene.2021.740216
- 65Brown LC, Stanton JD, Bharthi K, et al. Pharmacogenomic Testing and Depressive Symptom Remission: A Systematic Review and <scp>Meta‐Analysis</scp> of Prospective, Controlled Clinical Trials. Clin Pharma and Therapeutics. 2022;112:1303–17. doi:10.1002/cpt.2748
- 66Translate pharmacogenetic data into clinical action. Sequence2Script. 2023.https://www.sequence2script.com/#/ (accessed Jan 2023).
- 67Bousman CA, Wu P, Aitchison KJ, et al. Sequence2Script: A Web-Based Tool for Translation of Pharmacogenetic Data Into Evidence-Based Prescribing Recommendations. Front. Pharmacol. 2021;12. doi:10.3389/fphar.2021.636650
- 68Dunnenberger HM, Crews KR, Hoffman JM, et al. Preemptive Clinical Pharmacogenetics Implementation: Current Programs in Five US Medical Centers. Annu. Rev. Pharmacol. Toxicol. 2015;55:89–106. doi:10.1146/annurev-pharmtox-010814-124835
- 69Kimpton JE, Carey IM, Threapleton CJD, et al. Longitudinal exposure of English primary care patients to pharmacogenomic drugs: An analysis to inform design of pre‐emptive pharmacogenomic testing. Br J Clin Pharmacol. 2019;85:2734–46. doi:10.1111/bcp.14100
- 70Accelerating Genomic Medicine in the NHS. NHS England. 2022.https://www.england.nhs.uk/publication/accelerating-genomic-medicine-in-the-nhs/ (accessed Jan 2023).
- 71van der Wouden C, Cambon-Thomsen A, Cecchin E, et al. Implementing Pharmacogenomics in Europe: Design and Implementation Strategy of the Ubiquitous Pharmacogenomics Consortium. Clin. Pharmacol. Ther. 2017;101:341–58. doi:10.1002/cpt.602
- 72Chua EW, Foulds J, Miller AL, et al. Novel CYP2D6 and CYP2C19 variants identified in a patient with adverse reactions towards venlafaxine monotherapy and dual therapy with nortriptyline and fluoxetine. Pharmacogenetics and Genomics. 2013;23:494–7. doi:10.1097/fpc.0b013e328363688d
- 73McDermott JH, Leach M, Sen D, et al. The role of CYP2C19 genotyping to guide antiplatelet therapy following ischemic stroke or transient ischemic attack. Expert Review of Clinical Pharmacology. 2022;15:811–25. doi:10.1080/17512433.2022.2108401
- 74Rasool M, Malik A, Naseer MI, et al. The role of epigenetics in personalized medicine: challenges and opportunities. BMC Med Genomics. 2015;8. doi:10.1186/1755-8794-8-s1-s5
- 75Malki MA, Pearson ER. Drug–drug–gene interactions and adverse drug reactions. Pharmacogenomics J. 2019;20:355–66. doi:10.1038/s41397-019-0122-0
- 76Van Driest S, Shi Y, Bowton E, et al. Clinically Actionable Genotypes Among 10,000 Patients With Preemptive Pharmacogenomic Testing. Clin Pharmacol Ther. 2013;95:423–31. doi:10.1038/clpt.2013.229
- 77Youssef E, Kirkdale CL, Wright DJ, et al. Estimating the potential impact of implementing pre‐emptive pharmacogenetic testing in primary care across the UK. Br J Clin Pharmacol. 2021;87:2907–25. doi:10.1111/bcp.14704
- 78Weitzel KW, Cavallari LH, Lesko LJ. Preemptive Panel-Based Pharmacogenetic Testing: The Time is Now. Pharm Res. 2017;34:1551–5. doi:10.1007/s11095-017-2163-x
- 79Hicks J, Bishop J, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline forCYP2D6andCYP2C19Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin. Pharmacol. Ther. 2015;98:127–34. doi:10.1002/cpt.147
- 80Hicks J, Sangkuhl K, Swen J, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin. Pharmacol. Ther. 2017;102:37–44. doi:10.1002/cpt.597
- 81Greden JF, Parikh SV, Rothschild AJ, et al. Impact of pharmacogenomics on clinical outcomes in major depressive disorder in the GUIDED trial: A large, patient- and rater-blinded, randomized, controlled study. Journal of Psychiatric Research. 2019;111:59–67. doi:10.1016/j.jpsychires.2019.01.003
- 82Oslin DW, Lynch KG, Shih M-C, et al. Effect of Pharmacogenomic Testing for Drug-Gene Interactions on Medication Selection and Remission of Symptoms in Major Depressive Disorder. JAMA. 2022;328:151. doi:10.1001/jama.2022.9805
- 83Ontario Health (Quality). Multi-gene Pharmacogenomic Testing That Includes Decision-Support Tools to Guide Medication Selection for Major Depression: A Health Technology Assessment. Ont Health Technol Assess Ser 2021;21:1–214.https://www.ncbi.nlm.nih.gov/pubmed/34484487
- 84Corponi F, Fabbri C, Serretti A. Pharmacogenetics and Depression: A Critical Perspective. Psychiatry Investig. 2019;16:645–53. doi:10.30773/pi.2019.06.16
- 85Ginsburg GS, Cavallari LH, Chakraborty H, et al. Establishing the value of genomics in medicine: the IGNITE Pragmatic Trials Network. Genetics in Medicine. 2021;23:1185–91. doi:10.1038/s41436-021-01118-9
- 86IsHak WW, Mirocha J, Pi S, et al. Patient-reported outcomes before and after treatment of major depressive disorder. Dialogues in Clinical Neuroscience. 2014;16:171–83. doi:10.31887/dcns.2014.16.2/rcohen
- 87Islam F, Hain D, Lewis D, et al. Pharmacogenomics of Clozapine-induced agranulocytosis: a systematic review and meta-analysis. Pharmacogenomics J. 2022;22:230–40. doi:10.1038/s41397-022-00281-9
- 88Legge SE, Walters JT. Genetics of clozapine-associated neutropenia: recent advances, challenges and future perspective. Pharmacogenomics. 2019;20:279–90. doi:10.2217/pgs-2018-0188
- 89Legge SE, Pardiñas AF, Helthuis M, et al. A genome-wide association study in individuals of African ancestry reveals the importance of the Duffy-null genotype in the assessment of clozapine-related neutropenia. Mol Psychiatry. 2019;24:328–37. doi:10.1038/s41380-018-0335-7
- 90Patsopoulos NA, Ntzani EE, Zintzaras E, et al. CYP2D6 polymorphisms and the risk of tardive dyskinesia in schizophrenia: a meta-analysis. Pharmacogenetics and Genomics. 2005;15:151–8. doi:10.1097/01213011-200503000-00003
- 91Tiwari AK, Deshpande SN, Rao AR, et al. Genetic susceptibility to tardive dyskinesia in chronic schizophrenia subjects: III. Lack of association of CYP3A4 and CYP2D6 gene polymorphisms. Schizophrenia Research. 2005;75:21–6. doi:10.1016/j.schres.2004.12.011
- 92Liou Y-J, Wang Y-C, Bai Y-M, et al. Cytochrome P-450 2D6*10 C188T Polymorphism Is Associated with Antipsychotic-Induced Persistent Tardive Dyskinesia in Chinese Schizophrenic Patients. Neuropsychobiology. 2004;49:167–73. doi:10.1159/000077360
- 93de Leon J, Susce MT, Pan R-M, et al. Polymorphic Variations in GSTM1, GSTT1, PgP, CYP2D6, CYP3A5, and Dopamine D2 and D3 Receptors and Their Association With Tardive Dyskinesia in Severe Mental Illness. Journal of Clinical Psychopharmacology. 2005;25:448–56. doi:10.1097/01.jcp.0000177546.34799.af
- 94Fleeman N, McLeod C, Bagust A, et al. The clinical effectiveness and cost-effectiveness of testing for cytochrome P450 polymorphisms in patients with schizophrenia treated with antipsychotics: a systematic review and economic evaluation. Health Technol Assess. 2010;14. doi:10.3310/hta14030
- 95Lim K, Lam M, Zai C, et al. Genome wide study of tardive dyskinesia in schizophrenia. Transl Psychiatry. 2021;11. doi:10.1038/s41398-021-01471-y
- 96Drug-gene interaction testing may be about to go mainstream. Pharmaceutical Journal. 2022. doi:10.1211/pj.2022.1.162060
- 97Swen JJ, Nijenhuis M, de Boer A, et al. Pharmacogenetics: From Bench to Byte— An Update of Guidelines. Clin Pharmacol Ther. 2011;89:662–73. doi:10.1038/clpt.2011.34
- 98Picard N, Boyer J-C, Etienne-Grimaldi M-C, et al. Pharmacogenetics-based personalized therapy: Levels of evidence and recommendations from the French Network of Pharmacogenetics (RNPGx). Therapies. 2017;72:185–92. doi:10.1016/j.therap.2016.09.014
- 99Ross CJD, Visscher H, Sistonen J, et al. The Canadian Pharmacogenomics Network for Drug Safety: A Model for Safety Pharmacology. Thyroid. 2010;20:681–7. doi:10.1089/thy.2010.1642
- 100Caudle K, Klein T, Hoffman J, et al. Incorporation of Pharmacogenomics into Routine Clinical Practice: the Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline Development Process. CDM. 2014;15:209–17. doi:10.2174/1389200215666140130124910
- 101Abdullah-Koolmees H, van Keulen AM, Nijenhuis M, et al. Pharmacogenetics Guidelines: Overview and Comparison of the DPWG, CPIC, CPNDS, and RNPGx Guidelines. Front. Pharmacol. 2021;11. doi:10.3389/fphar.2020.595219
- 102Barbarino JM, Whirl‐Carrillo M, Altman RB, et al. PharmGKB: A worldwide resource for pharmacogenomic information. WIREs Mechanisms of Disease. 2018;10. doi:10.1002/wsbm.1417
- 103Table of Pharmacogenetic Associations. FDA. 2022.https://www.fda.gov/medical-devices/precision-medicine/table-pharmacogenetic-associations (accessed Jan 2023).
- 104Guideline on good pharmacogenomic practice. European Medicines Agency. 2018.https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-good-pharmacogenomic-practice-first-version_en.pdf (accessed Jan 2023).
- 105Drugs and health products. Government of Canada. 2022.https://www.canada.ca/en/health-canada/services/drugs-health-products.html (accessed Jan 2023).
- 106Clinical Commissioning Urgent Policy Statement: Pharmacogenomic testing for DPYD polymorphisms with fluoropyrimidine therapies. NHS England. 2020.https://www.england.nhs.uk/publication/clinical-commissioning-urgent-policy-statement-pharmacogenomic-testing-for-dpyd-polymorphisms-with-fluoropyrimidine-therapies/ (accessed Jan 2023).
- 107Personalised prescribing: using pharmacogenomics to improve patient outcomes. Royal College of Physicians and British Pharmacological Society. 2022.https://www.rcp.ac.uk/projects/outputs/personalised-prescribing-using-pharmacogenomics-improve-patient-outcomes (accessed Jan 2023).
- 108Pharmacogenomics – the future of pharmacy. Royal Pharmaceutical Society. 2022.https://www.rpharms.com/development/pharmacogenomics (accessed Jan 2023).
- 1092022–2023 projects. NHS North West Genomic Medicine Service Alliance. 2022.https://www.nw-gmsa.nhs.uk/about-us/our-regional-and-national-projects/our-2022-2023-projects (accessed Jan 2023).
- 110Karamperis K, Koromina M, Papantoniou P, et al. Economic evaluation in psychiatric pharmacogenomics: a systematic review. Pharmacogenomics J. 2021;21:533–41. doi:10.1038/s41397-021-00249-1
- 111Rafi I, Crinson I, Dawes M, et al. The implementation of pharmacogenomics into UK general practice: a qualitative study exploring barriers, challenges and opportunities. J Community Genet. 2020;11:269–77. doi:10.1007/s12687-020-00468-2
- 112Maciel A, Cullors A, Lukowiak AA, et al. Estimating cost savings of pharmacogenetic testing for depression in real-world clinical settings. NDT. 2018;Volume 14:225–30. doi:10.2147/ndt.s145046
- 113Morris SA, Alsaidi AT, Verbyla A, et al. Cost Effectiveness of Pharmacogenetic Testing for Drugs with Clinical Pharmacogenetics Implementation Consortium ( <scp>CPIC</scp> ) Guidelines: A Systematic Review. Clin Pharma and Therapeutics. 2022;112:1318–28. doi:10.1002/cpt.2754
- 114Roden DM, McLeod HL, Relling MV, et al. Pharmacogenomics. The Lancet. 2019;394:521–32. doi:10.1016/s0140-6736(19)31276-0
- 115PREemptive Pharmacogenomic Testing for Preventing Adverse Drug REactions (PREPARE). clinicaltrials.gov. 2022.https://clinicaltrials.gov/ct2/show/NCT03093818
- 116Maruf AA, Bousman CA. Approaches and hurdles of implementing pharmacogenetic testing in the psychiatric clinic. PCN Reports. 2022;1. doi:10.1002/pcn5.26
- 117Pharmacogenomics: three challenges to the NHS. Genomics Education Programme. 2019.https://www.genomicseducation.hee.nhs.uk/blog/pharmacogenomics-three-challenges-to-the-nhs/ (accessed Jan 2023).
- 118Out of Area Placements in Mental Health Services. NHS Digital. 2023.https://digital.nhs.uk/data-and-information/publications/statistical/out-of-area-placements-in-mental-health-services#latest-statistics (accessed Jan 2023).
- 119Fan M, Bousman CA. Commercial Pharmacogenetic Tests in Psychiatry: Do they Facilitate the Implementation of Pharmacogenetic Dosing Guidelines? Pharmacopsychiatry. 2019;53:174–8. doi:10.1055/a-0863-4692
- 120Bousman CA, Jaksa P, Pantelis C. Systematic evaluation of commercial pharmacogenetic testing in psychiatry. Pharmacogenetics and Genomics. 2017;27:387–93. doi:10.1097/fpc.0000000000000303
- 121Wouden CH, van Rhenen MH, Jama WOM, et al. Development of the <scp>PG</scp> x‐Passport: A Panel of Actionable Germline Genetic Variants for Pre‐Emptive Pharmacogenetic Testing. Clin. Pharmacol. Ther. 2019;106:866–73. doi:10.1002/cpt.1489
- 122Fatumo S, Chikowore T, Choudhury A, et al. A roadmap to increase diversity in genomic studies. Nat Med. 2022;28:243–50. doi:10.1038/s41591-021-01672-4
- 123Magavern EF, Gurdasani D, Ng FL, et al. Health equality, race and pharmacogenomics. Brit J Clinical Pharma. 2021;88:27–33. doi:10.1111/bcp.14983
- 124Constantinescu A-E, Mitchell RE, Zheng J, et al. A framework for research into continental ancestry groups of the UK Biobank. Hum Genomics. 2022;16. doi:10.1186/s40246-022-00380-5
- 125Liko I, Lai E, Griffin RJ, et al. Patients’ Perspectives on Psychiatric Pharmacogenetic Testing. Pharmacopsychiatry. 2020;53:256–61. doi:10.1055/a-1183-5029
- 126McCarthy MJ, Chen Y, Demodena A, et al. Attitudes on pharmacogenetic testing in psychiatric patients with treatment‐resistant depression. Depression and Anxiety. 2020;37:842–50. doi:10.1002/da.23074
- 127Kastrinos A, Campbell-Salome G, Shelton S, et al. PGx in psychiatry: Patients’ knowledge, interest, and uncertainty management preferences in the context of pharmacogenomic testing. Patient Education and Counseling. 2021;104:732–8. doi:10.1016/j.pec.2020.12.021
- 128Holden C, Bignell L, Mukhopadhyay S, et al. The public perception of the facilitators and barriers to implementing personalized medicine: a systematic review. Personalized Medicine. 2019;16:409–20. doi:10.2217/pme-2018-0151
- 129Asiedu GB, Finney Rutten LJ, Agunwamba A, et al. An assessment of patient perspectives on pharmacogenomics educational materials. Pharmacogenomics. 2020;21:347–58. doi:10.2217/pgs-2019-0175
- 130Shishko I, Almeida K, Silvia RJ, et al. Psychiatric pharmacists’ perception on the use of pharmacogenomic testing in the mental health population. Pharmacogenomics. 2015;16:949–58. doi:10.2217/pgs.15.22
- 131Chan CYW, Chua BY, Subramaniam M, et al. Clinicians’ perceptions of pharmacogenomics use in psychiatry. Pharmacogenomics. 2017;18:531–8. doi:10.2217/pgs-2016-0164
- 132Vest BM, Wray LO, Brady LA, et al. Primary care and mental health providers’ perceptions of implementation of pharmacogenetics testing for depression prescribing. BMC Psychiatry. 2020;20. doi:10.1186/s12888-020-02919-z
- 133Laplace B, Calvet B, Lacroix A, et al. Acceptability of Pharmacogenetic Testing among French Psychiatrists, a National Survey. JPM. 2021;11:446. doi:10.3390/jpm11060446
- 134Personalised Medicine Approach For Fluoropyrimidine-based Therapies. UK Chemotherapy Board. 2020.https://4bd2316d-e45d-4e90-96b5-431f1c12dd3e.filesusr.com/ugd/638ee8_852a3c89120242118e76d8cc303b2fef.pdf (accessed Jan 2023).