Abstract
A large proportion of the current pharmacopoeia can trace its origin back to nature. Biodiversity in the Earth’s largest habitat, the ocean, is greater than that on land, suggesting a huge potential for new bioactive chemistry. Compounds isolated from marine organisms have novel structures and modulate human disease targets with novel mechanisms of action. Around 15–20 marine-derived compounds have been approved for clinical use against cancer, pain, viral infection and heart disease. Some of the most potent compounds are directed towards cancer cells via linkage to an antibody. The approved marine-derived pharmaceuticals have been produced via (semi)-synthesis, while expression of the biosynthetic gene clusters in a microbial host is a future alternative for the production of those compounds. Technical improvements in the extraction, isolation and structural characterisation of marine-derived compounds, as well as the reduction in effort wasted in isolation of known compounds, have sped up the discovery process, meaning that industry is taking a renewed interest in this field as a source of pharmaceuticals with novel mechanisms of action to treat human disease with unmet need.
Key words: Marine natural products, drug discovery, aquaculture, semi-synthesis antibody-drug conjugates
Introduction
Compounds of natural origin constitute a large proportion of the current pharmacopoeia, with around 70% of currently used antibiotics and anticancer agents tracing back to natural sources, such as plants and microbes1. While plant-based medicines have a recorded history of over 5,000 years and microbially-derived medicines have been in use for almost a century, compounds from marine sources have recently only been investigated as potential medicines comparatively recently1. Recent clinical approvals started at the turn of the millennium and to date more than 15–20 marine-derived pharmaceuticals have been clinically approved, mainly for cancer treatment predominantly owing to funding by the US National Cancer Institute from the 1960s2.
The exact number of pharmaceuticals of marine origin that have received approval is debated, as some are closely related structurally2. This review outlines why marine natural products (MNPs) are an attractive prospect for future drug discovery, provides a history of the field, successes in the clinic and challenges associated with gaining clinical approval, including the need to obtain sufficient amounts of often very complex compounds. The pipeline of compounds currently in preclinical evaluation is promising and sophisticated technologies will improve the ability to discover, isolate, identify and modify marine-derived compounds with potent and selective activity, and new mechanisms of action against a range of human diseases that are currently without adequate treatment.
Marine biodiversity as sources of new pharmaceuticals
The driver behind natural product (NP) discovery for pharmaceutical applications is that the greater the biological diversity investigated, the greater the chemical diversity will be found. For this reason, the pioneers of MNP research investigated marine invertebrates from phyla with no terrestrial representatives, such as sponges, soft corals and sea mosses3. Marine animal biodiversity at the level of phylum is greater than that on land; while there are around 20 marine phyla with no terrestrial representatives, there are few animal phyla that have only terrestrial representatives4.
Global distribution can be mapped to show that there are biodiversity hotspots and that the deep seas have only been sparsely investigated5. This phyletic diversity at a very high level is complemented by a high diversity of marine microorganisms, the extent of which is not yet fully appreciated. It is hypothesised that conditions in the seas increase evolutionary pressure to produce unique bioactive molecules6. These conditions include high hydrostatic pressure, high salt concentrations, low light and oxygen levels, as well as the fact that many invertebrates have no obvious physical defences and lack an immune system7. Filter-feeding invertebrates should be subject to predation and overgrowth as well as infection by pathogenic microorganisms, but some have developed a chemical defence strategy that acts as an alternative to an immune system. The NPs produced as part of this chemical defence often have properties similar to drug molecules as they have to traverse many of the same barriers to reach the site of action, which may be an intracellular receptor6.
Once screening of marine invertebrates started, it appeared that certain phyla had a higher occurrence of bioactivity against human disease screens than terrestrial NPs. Success rates are hard to quantify, but a simplistic estimation divides the number of known MNPs (around 40,000) by the number of approved marine-derived drugs (between 15 and 20), giving a success rate of around 1 in 2,000–2,7008. MNPs appeared to have greater chemical diversity and a greater frequency of new skeletons than other natural sources, suggesting they could have novel bioactivities and modes of action against disease targets9. A recent study demonstrated that three-quarters of marine microbial NPs are structurally closely related to those from terrestrial microbes10. The occurrence of many of these unique microbial NPs in marine invertebrates suggests the potential microbial origin of some marine invertebrate NPs. Some of these unique skeletons have shown unique modes of action, such as ecteinascidin-743, isolated from a Caribbean sea squirt (see below), which binds to the DNA minor groove, thus bending the helix to the major groove. This triggers a cascade of events that affects several transcription factors, DNA binding proteins and DNA repair pathways, resulting in perturbation of the cell cycle11. There are many other examples of novel modes of activity that will be discussed throughout this article. The current success of MNPs in the clinic and the pipeline appears to be linked to the evolutionary success of marine invertebrates and micro-organisms, as well as the structurally unique chemistry they produce.
Processes for marine natural product drug discovery
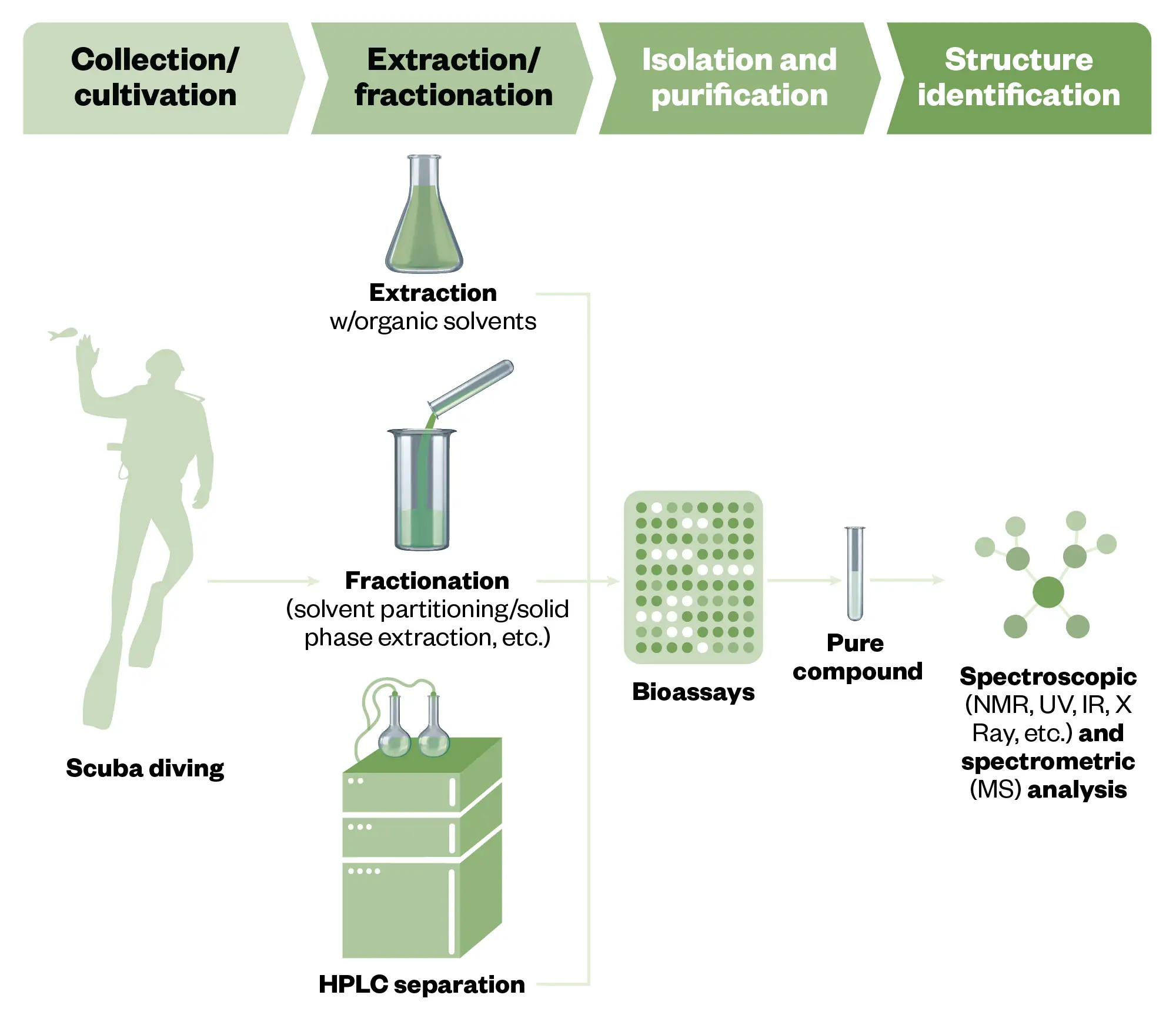
A typical workflow for MNP drug discovery starts by obtaining biomass; for instance, a marine invertebrate or a microbial culture obtained from a marine source (e.g. sediment, sponge etc), which is then subjected to large-scale fermentation (see Figure 1).
The Pharmaceutical Journal/Shutterstock.com
This biomass is then extracted using chemical solvents, after which the solvent is removed generating an extract potentially containing hundreds to thousands of compounds. Downstream steps are to fractionate these extracts to remove water-soluble compounds, such as salts, sugars and highly soluble metabolites and lipophilic compounds (e.g. fats). The fractions can be subjected to biological assays relevant to human disease followed by a cycle of chromatographic purification and assays to pinpoint the biologically active compounds12. Often these steps are complemented by using liquid chromatography — mass spectrometry (LC-MS) to determine which of these extracts and fractions contain compounds that have not previously been discovered to target effort towards novel bioactives. Once a pure bioactive compound is identified, its structure is determined using MS in conjunction with nuclear magnetic resonance (NMR) data13. The pure compound can either be used as is, or its structure modified. In all cases, the material will need to be obtained in higher quantities for the downstream mechanism of action (MoA) and animal testing before embarking on an investigational new drug application, potentially leading to human clinical trials, which can take many years before approval. More information can be found under ’Supplying enough material for clinical trials and clinical use’2.
Investigating natural products from marine sources
NPs have been utilised as ‘drug entities’ for millennia, including morphine and, more recently, penicillin. Towards the end of the 20th century, there was a significant move towards “high technology systems” including the concept of diversity-oriented syntheses (DOS), whereby millions of compounds could be easily synthesised and then tested in advanced biological systems. As of October 2019, after testing tens of millions of compounds in countless screening systems, at best four approved drugs (meaning approved by the US Food and Drug Administration (FDA) and/or the corresponding entities globally), have been identified for any human disease from utilising such methodologies1. This demonstrates that such systems are useful for developing an active agent once identified, but they fail in the discovery phase.
With the advent of new analytical systems that permit the identification of relationships between NP isolates, coupled with the application of new genomic analytical capabilities, particularly with microbial sources, these capabilities have enabled identification of many potential new leads in disease-related screening systems (see below)1.
Back to the 1950s and early marine investigations
Although there were chemists who liked to dive (mainly in warm waters) in the late 1950s, in the early 1960s and later, disparate opportunities came together. The first was the availability of reliable commercial scuba gear permitting non-military divers to explore up to roughly 40 metres deep using compressed air and then with mixed gases to greater depths. Early funding (1940s) possibly by the US (NIH) for basic studies on sponge metabolites led to some interesting compounds that can be considered the first marine bioactive agents that led to drugs used in humans. Funding from the US National Cancer Institute (NCI) in the 1960s to specific groups to collect, fractionate and then have in vitro/in vivo assays performed by NCI contractors. These groups worked on plants (USDA and Wall) and marine invertebrates (Pettit and Rinehart). Later funding (1986 onwards) for collections via NCI contractors and subsequent testing led to later drug entities (see below).
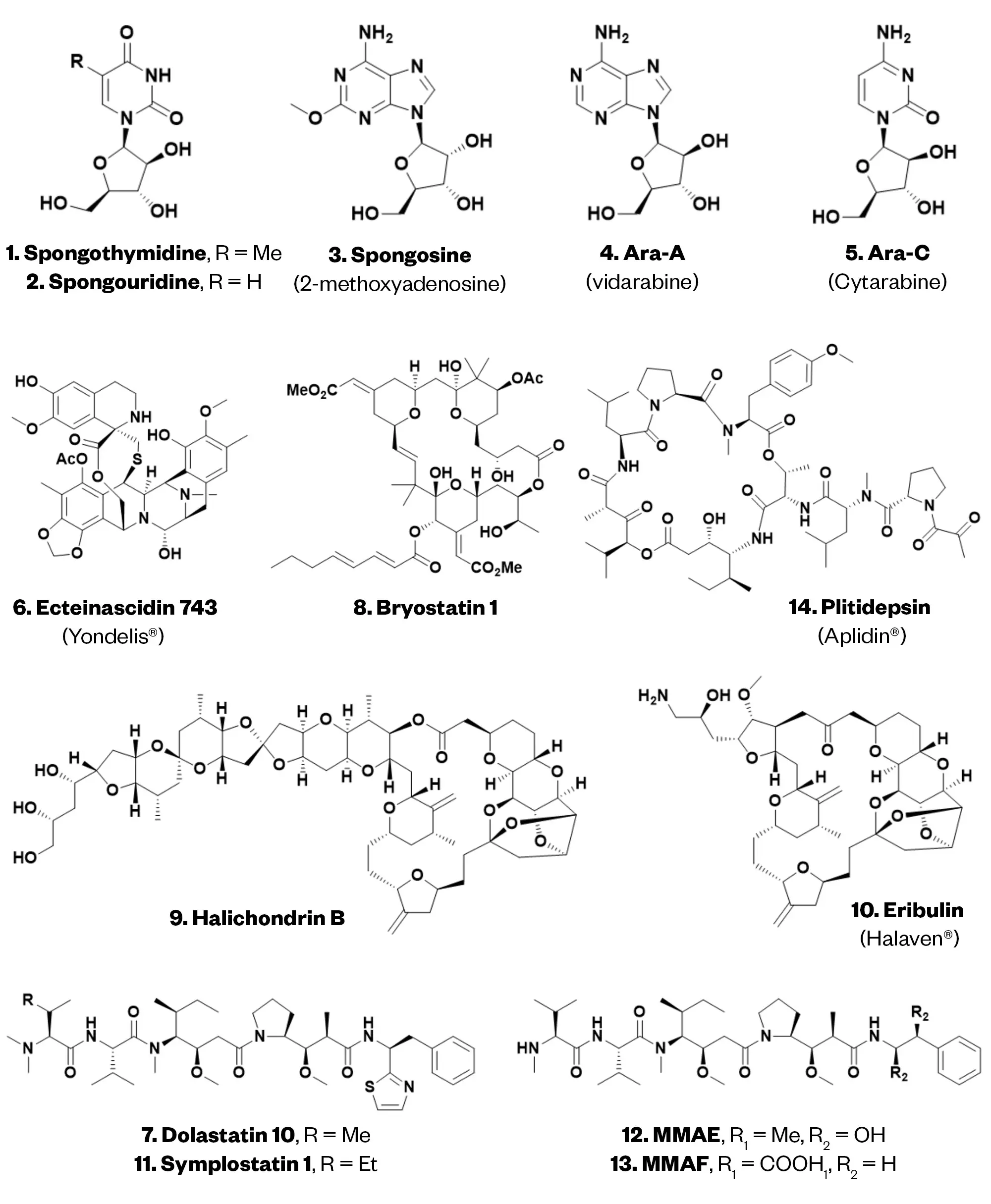
From the middle of the 1940s onwards, Bergmann and Feeney investigated the Caribbean sponge, (Tethya crypta) for novel terpenes, and from acetone extracts they discovered novel nucleosides with arabinose as the sugar moiety. The group reported the isolation and identification of spongothymidine (compound 1), spongouridine (compound 2) and spongosine (2-methoxyadenosine; compound 3), from the sponge (now renamed Cryptothethya crypta)14–16. Although none were developed as drugs, they led to a significant number of both antiviral and antitumour leads/drugs over the next few years, such as acyclovir against herpes virus or 6-mercaptopurine, the nucleoside-based anticancer drug17.
The early reports of potential bioactivity from compounds 1–3 concerned their potential as sources of uracil for E. coli auxotrophs18. Research by synthetic chemists took the information from the first Bergmann paper and synthesised spongouridine (compound 2)19; other arabino-nucleosides in 1957 by Fox et al.20 and Walwick’s synthesis in 1959 of Ara-C (compound 5)21 provided the necessary material for Lee et al. to uncover the antitumour potential of the semisynthetic derivative Ara-A in 1960 (compound 4)22 and for Evans et al. subsequent discovery of the utility of Ara-C (compound 5) as an antitumour agent in 196123.
In 1969, microbiologists at Parke-Davis demonstrated the fermentative production, rather than semisynthesis, of Ara-A using the terrestrial microbe Streptomyces antibioticus. This was only published as a patent by Parke-Davis24. Fifteen years later, the Cimino group in Italy reported the isolation of “biosynthesised Ara-A” together with spongouridine (compound 2) from the Mediterranean gorgonian coral Eunicella cavolini25. In this case, synthetic chemists had made the compound before it was discovered in nature. These discoveries implied a microbial source, even though the “nominal sources” ranged from invertebrates to terrestrial microbes.
In 2015, the Gerwick group used a fresh collection of the Caribbean sponge Bergmann used and isolated a strain of Vibrio harveyi from it; on fermentation it yielded spongosine (compound 3). Therefore, it is a reasonable assumption that bacteria were responsible for the Bergmann compounds 50 or more years earlier26.
Microbial involvement in generation of marine natural products
Moving into the 21st century, it has become increasingly obvious that microbes, whether they are currently fermentable, are most likely the actual source(s) of bioactive materials isolated from the Porifera and other marine invertebrates. The Piel group at ETH Zurich and their collaborators working with a sponge originally from Palau, and with similar sponges from Okinawa, have demonstrated that the source of the majority (>20 of the 35 plus) of bioactive agents isolated from the Palauan sponge are produced by a yet unfermented bacterium27.
The T. swinhoei sponges, collected in Palau and Japan, both produce an analogue of the Amazonian Paederus beetle toxin (itself produced by an as-yet uncultivated Pseudomonad on the female beetle), demonstrating that nature utilises similar genomes across the gamut of sources28 (see Figure 2). In 2022, a review article by Hemmerling and Piel highlighted how the biosynthetic novelty in bacterial genomes can be accessed, and how to further develop access to genomes29. To date, there has not been an unequivocal isolation of a completely sponge-sourced bioactive agent, despite searching for such an agent for over 60 years.
The Pharmaceutical Journal
Two other early bioactive agents in the first case (the dolastatins) are now definitively from a prokaryotic cyanobacterium, even though they were first isolated from the sea slug Dolabella auricularia (see Figure 3). The second, the multiple ecteinascidins, date from studies in the Rinehart laboratory in the early 1980s onwards originally from the marine tunicate Ecteinascidia turbinata (compound 6; see Figure 2). In 2011, the Sherman group demonstrated that the actual producer in the tunicate was a microbe (Candidatus Endoecteinascidia frumentensis) symbiotic with the tunicate30. In 2015, it was shown that the bacterium was, in fact, a “stripped” genomic version that in exchange for protection from predators, the tunicate provided essential nutrients to the microbe31. The fact the bacterial genome was “stripped” indicates that the symbiosis between it and the host had been established a long time ago.
Supplying enough material for clinical trials and clinical use
Marine (micro)organisms are characterised by an impressive chemical diversity and a thorough analysis of a single marine invertebrate can afford tens, if not hundreds, of secondary metabolites that can be used to build a “natural combinatorial library” for drug development32. Often these metabolites are rare owing to being contained at low levels in the living organisms, which are, in turn, available only in a few specimens because their collection is limited by environmental concerns. Progress in structure elucidation techniques now allows full NMR and MS spectral analysis with less than a 1 milligram sample, but the preliminary in vivo pharmacological screening and further drug development need larger amounts of test compounds13.
The scientific literature is full of cases of marine metabolites that have shown potent and promising bioactivities during preliminary screening, but whose limited availability has hampered further development. Progress in aquaculture and chemical synthesis or semi-synthesis have provided valid alternatives to solve the supply bottleneck. Recent advances in molecular genetics allow the identification of biosynthetic genes in producing organisms and their cloning into microbes suitable for large-scale fermentation. This approach is promising, but has only proved successful at the research level and not yet at the industrial scale for marketable marine drugs. Developments in this area will be covered in more detail in a later section1.
The following examples of aquaculture and chemical synthesis or semi-synthesis illustrate how the discovery of potential leads from diverse marine sources has been translated into drugs that have entered the market, by allowing large-scale production. These successful examples ensure that pharmaceutical companies now regard marine natural product-derived pharmaceuticals with renewed interest.
Aquaculture
Many pharmaceutically interesting marine macro- or microorganisms are difficult to culture under artificial conditions33. A detailed understanding of living conditions in the natural environment is necessary to develop the optimal cultivation methods and, in the case of microorganisms, co-cultivation of two or more different strains, also called mixed fermentation, has provided a significant enhancement in the production of constitutive compounds or accumulate cryptic ones34.
In the case of macroorganisms, aquaculture, namely the controlled cultivation of aquatic organisms in the sea or in artificial environments, has proved to be a valid option for large-scale production in several cases. One of the best examples of successful aquaculture supply is that of bryostatin 1 (compound 8), a macrocyclic polyketide lactone from the bryozoan Bugula neritina, that acts as a potent modulator of protein kinase C and has been in clinical trials as an anti-cancer agent, an anti-AIDS/HIV agent and for treatment of patients with Alzheimer’s disease35,36. The initial preclinical and clinical tests on bryostatin 1 required gram quantities of the compound obtained by collecting 13 metric tonnes of wet Bugula neritina35. This strategy could not sustain further experiments, and possible clinical use of the compound, so scientists resorted to aquaculture. The producing bryozoan was successfully grown using both in-sea and on-land aquaculture and the levels of bryostatin were high enough to obtain the needed supply of the compound37. The producing bryozoan was successfully grown using a combination of in-sea and on-land aquaculture that involved the settlement of “tadpole” larvae on artificial materials, followed by a pre-grow period in tanks, and then grow-out in the ocean on anchored PVC pipe structures. The levels of bryostatin were high enough to obtain the needed supply of the compound37.
Chemical synthesis or semi-synthesis
Modern synthetic chemists can build extremely complex molecules with clinical potential that are identical to or based on the NP structure, even in the case of stereochemically dense molecules. For example, halichondrin B (compound 9) is a complex polycyclic polyketide, first reported from the sponge Halichondria okadai, which was collected in Japan, that acts as antimitotic by inhibiting microtubule dynamics, through binding to high-affinity sites at the positive ends of existing microtubules38. Similarly to bryostatin 1 (compound 8), early preclinical investigations on halichondrin B (compound 9) were made possible only by a large campaign of sponge collection conducted after an environmental impact assessment. In parallel to some successful attempts of aquaculture of the sponge, Kishi at Harvard University and the Japanese company Eisai were involved in the total synthesis of the natural halichondrin B (compound 9) reported in 2019 and earlier in structural simplifications, with replacement of the macrocycle with a ketone group and truncation of the tail region of the molecule39,40. It was discovered that the synthetically manipulated molecule, named eribulin (compound 10), was even more active in vivo and had a better therapeutic index than halichondrin B. Eribulin mesylate is an anticancer medication used to treat refractory metastatic breast cancer and liposarcoma41.
Another way to conveniently apply advanced organic synthesis principles to the production of drugs is semisynthetic transformations on MNPs. In this case, easily available compounds are converted by chemical processes into the desired product. An example of this is ecteinascidin 743 (compound 6), a complex alkaloid isolated from the tunicate Ecteinascidia turbinata that interferes with transcription42. Ecteinascidin’s preclinical and clinical trials to Phase II were carried out using bulk collection of the tunicate (around 100 tonnes of biomass). In this case, the solution toward large-scale production of the compound could not be total synthesis, as none of the published syntheses was amenable to this. The structural similarity of ecteinascidins with saframycins, known terrestrial microbial tetrahydroisoquinoline antitumour antibiotics, suggested the application of a semisynthetic strategy43. As a result, all the current supply of trabectedin (ecteinascidin 743), for the treatment of advanced soft-tissue sarcoma and ovarian cancer, is obtained by semi-synthesis from the microbial product cyanosafracin B, produced by large-scale fermentation of Pseudomonas fluorescens44.
Success in the clinic
As previously described, the first marine-derived leads to drugs for human use were the arabinose-linked nucleosides. The second compound family to receive serious attention was a series of linear peptides isolated by the Pettit group from the sea hare Dolabella auricularia. To contextualise the scale necessary at that time, the first three collections in the Indian Ocean beginning in 1965 were at the 2–100kg level and resulted in the discovery of several cytotoxic agents following bioactivity-driven isolation against murine P388 leukaemia45–47. Finally, in 1987, the Pettit group published the formal structure of dolastatin 10 (compound 7), following the collection of 1,600kg of material and subsequent isolation of 28.7mg of the product45. Owing to the lack of isolable material, dolastatin 10 was made available by total synthesis, with a US patent issued in 199046,47.
Subsequently, the “true source” of the dolastatins was discovered by natural product chemists associated with the Moore group at the University of Hawaii. Initially, symplostatin 1 (compound 11) isolated from Symploca hydnoides (a cyanobacterium) from Guam was a close chemical analogue of dolastatin 10 (compound 7)48. Three years later, Luesch et al. discovered dolastatin 10 from another collection of Symploca collected in Palau, later reclassifified as Caldora penicillata49,50.
The isolation of dolastatin 10 in 10,000-times higher yield than from the sea hare, coupled with ecological evidence with Dolabella auricularia being a generalist herbivore, indicated that cyanobacteria are the producer of this compound class and its source in the sea hare. Following the discovery from cyanobacteria from the Pacific Ocean, the Luesch and Paul groups found these compounds in marine cyanobacteria from Florida and the Caribbean, suggesting their ubiquitous presence in reefs across the globe and providing a robust and reliable compound source by recognising ecological relationships and biosynthetic signatures, as these modified peptides possess “classical” cyanobacterial structural motifs51. Recently, the Luesch group working with the Ding group at the University of Florida, published data on the biosynthetic gene cluster for dolastatin 10 from the producing cyanobacterium, thus effectively “completing the biosynthetic circle” for this compound52.
Dolastatin 10 (compound 7) possesses extraordinary broad-spectrum activity in the picomolar range against various cancer cell types (e.g. melanoma, colorectal, ovarian), it targets the protein tubulin, which is critically involved in cell proliferation52. Synthetic access to dolastatin 10 (compound 7) has provided an opportunity to synthesise strategic analogues, including monomethyl auristatins E and F (MMAE [compound 12] and MMAF [compound 13]), which possess altered permeability and diffusion properties53. Although dolastatin 10 (compound 7) and derivatives went into clinical trials as potential anticancer drugs, none of these “simple single compounds” went beyond phase II clinical trials owing to their innate toxicities54.
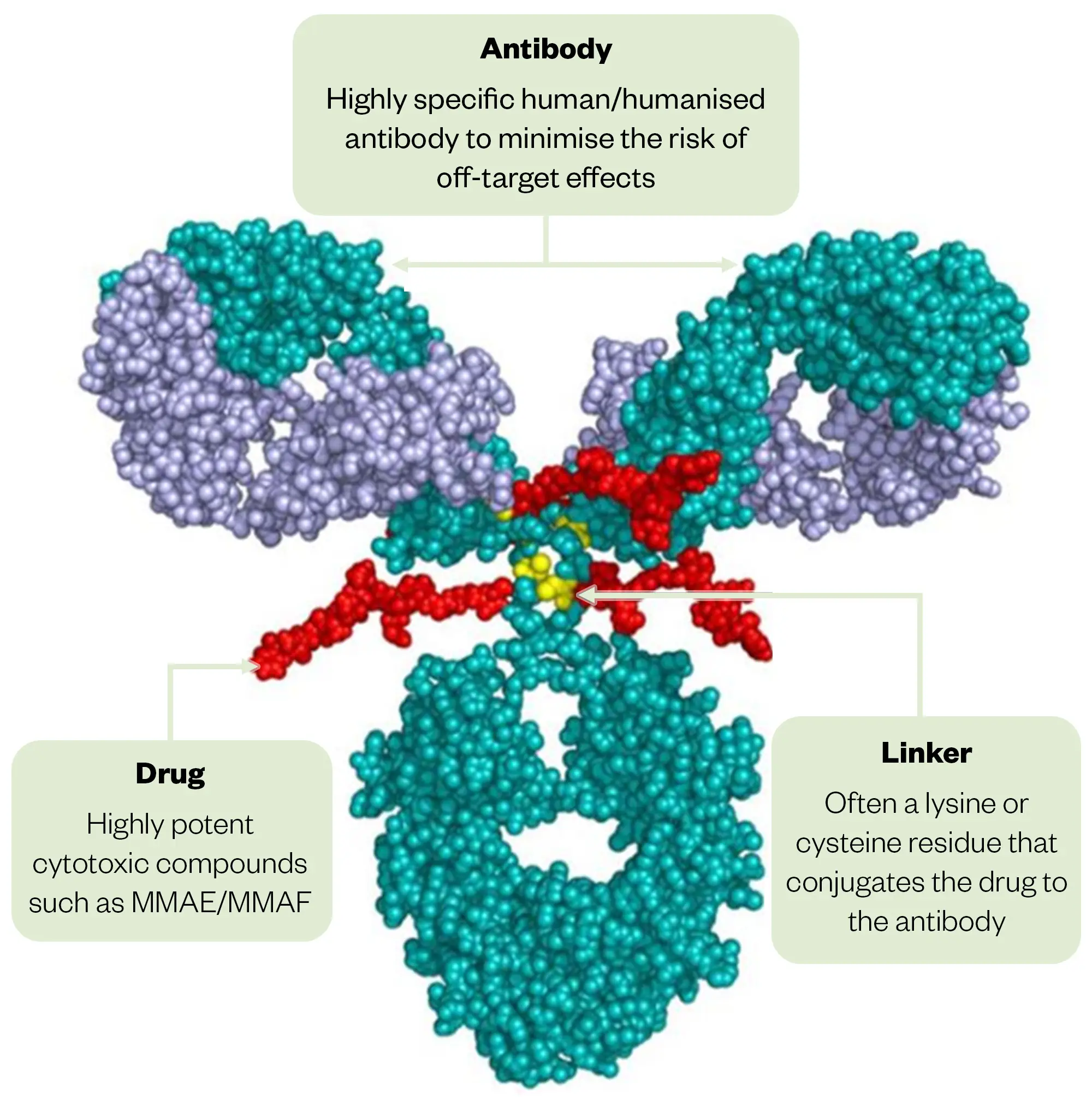
The advent of the antibody drug conjugate (ADC) platform as an enabling technology has been a game changer for MNP development; it has provided the opportunity to use antibodies to guide the cytotoxin to the cancer cell, circumventing toxicity owing to systemic exposure (see Figure 4). The toxic payload is then released to target the cancer cells specifically54. The finding that MMAE (compound 12) and MMAF (compound 13), which possess an N-monomethyl valine instead of the N,N-dimethyl valine in dolastatin 10 (compound 7) as the N-terminal residue, retained biological activity54, opened up opportunities for conjugation at this residue via the amine. There are now at least 15 ADCs approved by the FDA and/or comparable governmental bodies outside of the United States, including five to seven that carry MMAE or MMAF as their cytotoxic payload2.

The Pharmaceutical Journal/Sources within image
The breakthrough was the development of brentuximab vedotin, which was first approved by the FDA in 2011 for Hodgkin’s lymphoma and anaplastic large-cell lymphoma55. Brentuximab vedotin was approved by NICE for the treatment of CD30-positive Hodgkin lymphoma in 201756. Other MMAE- and MMAF-containing ADCs have been FDA approved, in 2019 polatuzumab vedotin for B-cell lymphoma and enfortumab vedotin for refractory bladder cancer57,58. Polatuzumab vedotin was also approved by NICE in 2020 for the treatment of B-cell lymphoma59. However, enfortumab vedotin does not have UK authorisation60. In 2020 belantamab mafadotin was approved for relapsed/refractory multiple myeloma; this was withdrawn in November of 2022 owing to not meeting the required level of PFS54. In 2021, tisotumab vedotin was approved for recurrent or metastatic cervical cancer54. Tisotumab vedotinis is not yet approved by the MHRA, but a NICE technology appraisal is expected61. Disitamab vedotin was approved by the Chinese Center for Drug Evaluation (CDE) of the National Medical Products Administration (NMPA) for HER2-positive late-stage breast cancer patients in 202154. With the proven success of ADCs, the “flood gate” is now wide open, with more than 100 ADCs containing dolastatin 10-based payload undergoing preclinical and clinical development for many different cancers; the favourable toxicity profiles directly correlated with clinical impact. This example highlights that one potent marine compound can provide the basis for more cancer drugs with tremendous clinical impact, benefitting a large patient population and saving thousands of lives54.
The Pharmaceutical Journal
There have been other examples of success before the vedotins, including for non-oncology indications. Ziconotide was approved in 2006 for chronic pain treatment and as a primary alternative to morphine, displaying up to 1,000 times greater analgesic potency as a calcium ion channel blocker62. However, its requirement for a complex method of delivery to the spine has limited its utility. Approved marine hypertriglyceridemia drugs include omega-3 fatty acid and esters (2001: omega-3-acid ethyl esters, 2012: 2014: omega-3-carboxylic acids), and ethyl eicosapentaenoic acid (2012), all derived from marine sources, such as fish or krill63–65.
The biggest impact of marine drugs has been seen in the oncology space, where the investments were highest. Trabectedin/ET-743 was first approved in Europe in 2007 for the treatment of advanced soft-tissue sarcoma and has a unique mechanism of action based on interaction with the minor groove of DNA, bending the helix to the major groove66. This binding to DNA triggers a cascade of events affecting several transcription factors, DNA binding proteins, and DNA repair pathways, resulting in perturbation of the cell cycle. Trabectedin was approved by NICE for the same indication in 200967. Eribulin mesylate, targeting tubulin in a manner distinct from that of dolastatin 10, was approved by the FDA in 2010 and NICE in 2011, as a third-line treatment for metastatic breast cancer68. Linking eribulin to antibodies is emerging as a successful strategy to improve the pharmacological and toxicity profile69. Plitidepsin (Aplidin) (compound 14) discovered from the sea squirt Aplidium albicans was approved in Australia in late 2018 for the treatment of relapsed and refractory multiple myeloma patients in combination with dexamethasone (see Figure 2)70. By targeting eukaryotic elongation factor 1A (eEF1A), it also possesses antiviral activity and underwent clinical trials as a host-directed antiviral for the treatment of SARS-CoV-2 infections71.
Feeding the marine pharmaceutical pipeline
As most marine-derived compounds in the clinic or under clinical development target cancer, it is important to highlight that ongoing global MNP preclinical pharmacological research encompasses diverse clinically relevant pharmacological activities. A series of comprehensive and systematic reviews from 1998 to 2021 (with the latest of these reviews covering 2019 to 2021 is in preparation with details below and in Table 1) offer insight into numerous MNP that have demonstrated other activities beyond cancer72–80. A group at Midwestern University curated a website that covers marine sources and their potential, which can be found here2. A novel approach that uses warheads based on auristatin is also being developed where in place of a standard mAb targeting moiety, several novel, synthetic, structurally constrained bicyclic peptides (“Bicycle”) have been created that target EphA2 (BT5528) and Nectin-4 (BT8009)53,81. Unlike the limitations of current mAb-containing ADCs, bicycle toxin conjugates (BTCs) have been designed to have short plasma half-lives where effects are balanced by rapid and efficient tumour penetration, coupled with high Cmax levels allowing these BTCs to release the cell penetrant payload (MMAE) killing the target cell and producing efficient bystander killing53,81,82. Since this novel warhead/targeting modality is in early clinical development, there are still substantial hurdles to overcome, including human pharmokinetics and pharmacodynamics, the unknown potential adverse events that may be associated with this novel targeting modality, and the eventual proof of efficacy in humans82.
Where the next generation of MNP pharmaceuticals will come from
Developments in isolation procedures, including rapid high-content screening methods to strengthen current bioassays, and more precise structural elucidation methods along with a reduction in wasted effort via the use of bioinformatic tools and ‘omics’ approaches will likely yield the next generation of MNP pharmaceuticals83. Future drug candidates with improved properties will be developed by solving sustainable supply issues, discovering their bioactivity, mode of action (MoA), and optimising their pharmacological properties by the generation of analogues84.
Exploration of new natural sources such as those coming from extreme or deep environments or bringing uncultivable microorganisms into culture will generate new MNPs as future drug candidates. This will require development in engineering to sample at great ocean depths and an improved understanding of microbial physiology to enable the unculturable to be cultured85,86.
As less than 1% of microbes can be cultivated, partly owing to the fact that conventional methods may enrich the growth of specific microorganisms while hindering the cultivation of others, extensive efforts have been made to solve this problem. New innovative culturing methods have been developed in the last few years87,88. One example is the isolation chip (iChip) method, where microbes are localised to small wells at the single-cell level before being inserted into in situ environments. This has been shown to be a promise-culturing strategy to cultivate bacterial species that are not able to be cultured as they do not grow in laboratory conditions89. An improvement of this method has been recently published where the iChip and petri dish plating methods to cultivate bacteria from soil contaminated with or without crude oil were used. iChip improved the richness and evenness of bacterial isolates in both the natural and contaminated soil samples90. These systems work well on land, though would be restricted to perhaps mud flats or equivalent with marine-derived microbes. This is because it is a terrestrial technology that needs to be buried in the soil or sediment.
The bioassay-guided isolation approach to obtain pure bioactive compounds from active marine organism extracts is still the most successful approach91 (see above and Figure 1). However, the major drawback of this strategy is the time spent on repeated isolation of known structures. The development of rapid dereplication processes aimed at the early identification of known compounds is a priority issue in MNP drug discovery. Subsequently, several dereplication approaches have been developed92. The most used strategy uses hyphenated analytical methodologies, which combine chromatographic separation techniques, predominantly LC, and spectrometric data, mainly MS in the high resolution (HRMS) mode and/or with ion fragmentation (MS/MS) along with ultraviolet (UV) and/or nuclear magnetic resonance (NMR) spectroscopic techniques. The obtained data are analysed using databases of natural products (e.g. MarinLit for MNP, Natural Product Atlas for microbial NP, CyanoMetDB for cyanobacteria NP) and in the case of LC/MS can be analysed using molecular networking (MN)93 (see Figure 5). This computational strategy was introduced in 2012 and organises the mass spectral data based on the similarity between precursor ions’ mass fragmentations allowing the organisation of extensive tandem mass data sets and the mapping of the chemical diversity in the sample. In this way, it is possible to discover molecular families and substructures in MS/MS data. The combination of molecular networking with Global Natural Products Social Networking (GNPS), an open platform that compiles many metabolomic datasets is an effective approach for early identification of NP and annotation of their analogues without the need for isotopic labelling93,94.
More recently, feature-based molecular networking (FBMN) was created, showing more representative results by pre-processing the MS raw data with different computational tools95. The software detects the features of the precursor ions and filters the data by isotope patterns, retention time, or even ion mobility separation. These tools generate feature tables that can be submitted to GNPS obtaining much cleaner and more sensible results. GNPS includes more than 80,000 MS/MS spectra and is the most powerful informatics tool in NP dereplication and the most utilised among the NP research community94. The reference dataset is not complete but is improving continuously via the support of the natural products community (see Figure 5).
NMR based dereplication methods such as MixONat algorithm in Python for 13C NMR and computer-aided 13C NMR, represent a good alternative to the MS dereplication96. Although methods for dereplication using MS are more developed than for NMR, further research on NMR methods, especially when integrated with MS/MS methods, would increase dereplication efficiency and accuracy.
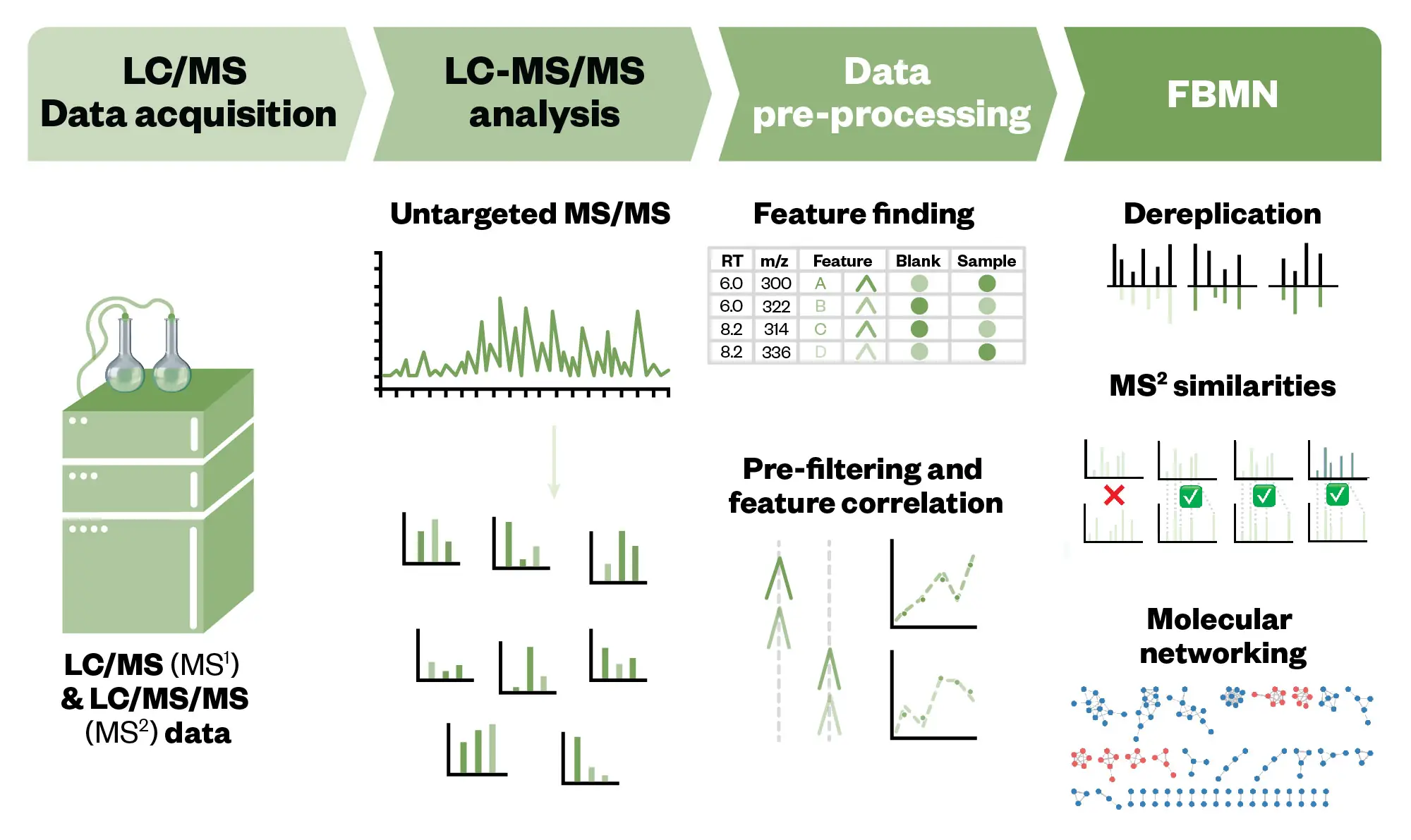
The Pharmaceutical Journal/Shutterstock.com
High throughput screening (HTS) continues to be a key strategy for identifying chemical compounds capable of modulating specific disease-related targets. New assays are constantly being developed to support drug discovery efforts and miniaturisation of assays allows biological screening to be performed with very small amounts of the pure NP or extracts. For example, a HTS was reported to identify compounds that inhibit biofilm formation using a Salmonella strain from 1,120 extracts97.
The reduction in the quantity needed for structural elucidation of complex natural products (i.e. the use microcryoprobe NMR) means only nanomoles of material are needed to solve a complete structure98.
Metabolomics can be defined as the qualitative and/or quantitative analysis of the whole set of low molecular weight metabolites present in a biological sample. Targeted metabolomics is used to analyse a set of defined metabolites and is commonly used to study human metabolism, whereas ‘untargeted metabolomics’ detects as many metabolites as possible, often without a complete identification of each, but allows clustering into groups99.
The application of ‘omics’ approaches, with the decrease in costs of DNA sequencing, the availability of MS (metabolomics, lipidomics, and proteomics) and the development of bioinformatics tools, have improved the analysis of biological systems and increased the possibilities of discovering new bioactive MNPs83.
MNPs are generated by biosynthetic enzymes that construct core scaffolds, introduce pharmacophores and create metabolic diversity through a complex series of enzymatic reactions and regulatory switches. These enzymes are encoded by a group of genes in close genomic proximity that are organised into biosynthetic gene clusters (BGC)100. As the structures of these enzymes are highly conserved, the BGCs can be used to computationally predict potential metabolites by genome mining (GM) which comprises computational methods for the automatic detection and annotation of BGCs from genomic data (see Figure 6). This approach allows for the prediction of the structure of the NP based on genetic information before its isolation and the structural elucidation along with their possible functional and chemical interactions. The low-cost and rapid development of genome sequencing methods with the development of bioinformatic analysis tools for BGC identification, and genome mining (GM) has become a key technology to exploit and explore NP diversity and is revolutionising the search for new MNPs101. Powerful computer analysis of genomes is allowing the identification of millions of putative biosynthetic pathways for novel molecules of possible avenues for creating entirely new drugs102. A plethora of new platforms and databases were developed to computationally mine genetic data such as the open-source platforms Antibiotics and Secondary Metabolite Analysis Shell (AntiSMASH), PRISM, BAGEL, TOUCAN, and others103. Once the BGC information is obtained, the next step is the prediction of NP structures. DeepRiPP, RiPPMiner, and RODEO (Rapid ORF Description and Evaluation Online) are some tools that allow the prediction of NP structures (see Figure 6)83.
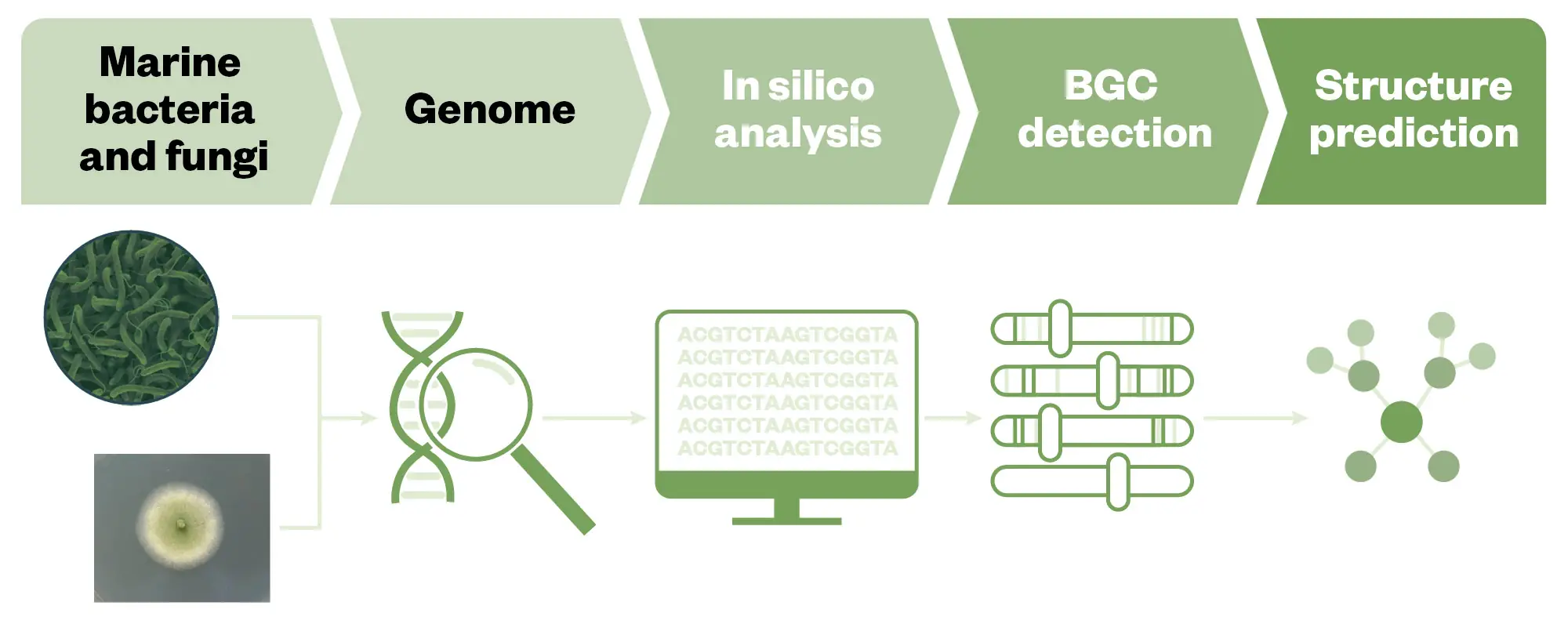
The Pharmaceutical Journal/PharmaMar
Another interesting tool is metagenomics, a process of extracting microbial genomes directly from environmental samples, regardless of sample type or microbial abundance. Metagenomics is a good alternative to overcome some of the difficulties in NP discovery by profiling microbial communities, such as the study of uncultivated species104. The metagenomic sequence data assembly of the endosymbiotic genus Entotheonella isolated from the marine sponge Theonella swinhoei allowed the identification of their ‘metabolic talent’ and led to the discovery of many unprecedented BGCs27.
Genetic engineering of different biosynthetic pathways requires different levels of engineering to manipulate them. The RiPP metabolites are the most straightforward and the manipulation of the BGCs or modifying enzymes can be achieved to generate new products either in host cells or via the application of modified enzymes on synthetic substrates105. Other BGCs such as non-ribosomal peptide synthetase (NRPS) or polyketide synthase (PKS) require more involved manipulation to generate new analogues of known compounds, although the use of mutations to allow non-natural substructures to be incorporated is very successful106. Future development in understanding of BGCs and genetic engineering will likely allow at first the generation of new analogues with improved properties and subsequently the generation of new chemical diversity to be tested against human disease.
The application of machine learning (ML) methods, defined as the study and application of algorithms that perform classification and prediction tasks through pattern recognition, to natural product research is accelerating the identification of new molecular entities for drug discovery. Prihoda et al. conducted a comprehensive review on ML methods in NP discovery with applications in genome mining, predictions of structures, and types of activities expected from those molecules107. More recently, a biosynthetic-based ML bioinformatic method to predict natural product biological activity from BGC sequences was published in 2021108. In this case, machine learning algorithms have been used to predict biological activity from biosynthetic gene clusters, enabling the identification of potential drug candidates from natural products. By analysing gene clusters, these methods help in understanding the biological activity of natural compounds and their potential as drug leads. Furthermore, a summary of the recent advancements in ML-assisted MS and NMR data analysis to establish the chemical structures of NPs was recently reported109. One example of machine learning-assisted structure annotation of natural products based on mass spectrometry (MS) and nuclear magnetic resonance (NMR) data is the use of tools like SMART 2.0, SMART-Miner, and COLMAR110–112. These tools utilise machine learning algorithms, such as convolutional neural networks and Bayesian approaches, to analyse complex NMR spectra and identify and annotate primary metabolites in natural product mixtures110–112. By leveraging AI techniques, researchers can enhance the efficiency and accuracy of structure elucidation and annotation of natural products, ultimately aiding in the discovery and characterisation of novel compounds from complex biological samples.
The output from GNPS can also be imported into other analysis programs such as Cytoscape, Metaboanalyst, QIIME 2 which offer ML tools. XGBoost classifier is an 1D 13C NMR data and machine learning method that predicts the chemical class of a NP with higher accuracy, outperforming other algorithms of the same type113. Computational methods to predict the MS of small molecules can be achieved with the use of machine learning (ISIS, CSI:FingerID) and molecular properties of unknown molecules are predicted by comparison with fragmentation trees using machine learning techniques83.
Artificial Intelligence (AI) is becoming a powerful tool for drug discovery, allowing scientists to predict how a natural product’s chemical structure affects its activity and which macromolecule might be its biological target. The applications of AI in natural product research, including the issues mentioned with ML, the pitfalls in training algorithms and approaches to avoid them were recently reviewed114. An example where AI was applied to drug discovery is the use of AlphaFold for predicting the 3D structure of proteins. AlphaFold is a deep learning system developed by DeepMind that can predict the 3D structure of proteins from their primary amino acid sequence by learning from a vast amount of protein structure data in the Protein Data Bank115. Another example is the deep learning techniques that have been applied to predict drug-target binding affinity, de novo drug-like molecule generation, property prediction, and protein pocket-conditioned molecular representations. These approaches leverage deep learning models to analyse molecular structures and properties, enhancing drug discovery processes115.
Application of these approaches to data derived from complex microbiomes, predicting structures from genomic signatures, and the types of activity we might expect from those molecules, enables us to better understand their “genomic language”102. For example, dereplication using integrated genome mining, MS/MS analysis along with in silico GNPS tools of the nearly complete genome of Streptomyces tendae VITAKN, which was isolated from the southern coast of India, yielded the discovery of columbamides, a new class of di- and trichlorinated acyl amides with cannabino-mimetic activity116. New peptides as members of the viequeamide class were discovered from a marine cyanobacterium by application of the Small Molecule Accurate Recognition Technology (SMART), which employs a convolutional neural network to classify HSQC spectra of organic molecules using pattern recognition principles117.
Future perspectives and conclusions
NPs are specialised metabolites, whilst most conform to small molecule drug parameters, many are larger and more complex than these ‘standard’ small molecules synthesised and screened by pharmaceutical companies against druggable targets. They represent privileged scaffolds as they have evolved to interact with biological targets. While MNPs have been successful in targeting validated disease-relevant proteins, one strength of MNPs includes the potential for discovering new mechanisms of action through phenotypic screening and subsequent validation of proteins that have not been successfully drugged previously or even considered to be targeted. MNPs are genetically encoded, providing opportunities for genomics-driven discovery approaches and synthetic biology to identify and optimise bioactive natural products50.
In that sense, MNPs will remain pioneering, lead to first-in-class pharmacological agents, and provide new starting points for the development of new pharmaceuticals. As demonstrated by dolastatin 10, while the target was not novel, the compound can be considered most-potent-in-class. Potency is a common theme among MNPs, which is one reason for their suitability as ADC payloads. We expect that the relevance of MNPs to the ADC field will further increase and that the ADC field will be dependent on MNPs to provide ultra-potent molecules that may be conjugated to cell- and target-seeking antibodies and other targeting agents such as the “bicyclic peptides”53,81.
Because of their larger size and complexity, MNPs occupy different chemical space than other compounds, which collectively allows for unusual interactions with protein complexes and for targeting protein-protein interactions, which are emerging as a new “target” class. Druggable space is evolving and previously “undruggable” targets or protein interfaces will increasingly be targeted by MNPs, therefore extending the druggable genome118. This can be extrapolated further to RNA-protein interactions, as shown for pateamine A by targeting eukaryotic initiation factor (eIF) 4A119. Therefore, it is conceivable that MNPs will target various disease-associated RNAs, which have been emerging as therapeutic targets and will lead to new classes of small-molecule pharmaceuticals, with RNA playing a major role beyond vaccines120.
In addition to acting at the level of nucleic acids (DNA, RNA), MNPs interact with membrane components or metabolites, such as glycans or lipids, and we will likely witness new target interactions to be deciphered beyond those in the standard repertoire. The field of marine natural products will continue to be characterised by high-level innovation through synergy with enabling technologies, such as ADC and cryo-electron microscopy.
The translation of NPs into drugs requires identification of the molecular targets, mechanism of action and even synthetic lethal vulnerabilities or pleiotropic effects where one gene influences two or more seemingly unrelated phenotypic characteristics of organisms. This is demonstrated by didemnin B, which inhibits protein elongation at the level of eukaryotic initiation factor eEF1A. It was shown to have potentially beneficial polypharmacology that can serve as a predictive pharmacogenomics response marker121. Identifying the most suitable therapeutic setting will be important for success. We expect to increasingly see repurposing of active compounds with the advance of new technologies and rigorous pharmacological assessment. New sensitive technologies to interrogate bioactivities at the molecular and cellular level will continue to be developed, increasing the value of these compounds with respect to their biomedical utility122.
The identification of mechanisms of action early in the discovery process will be key in accelerating their translation. A new target or mechanism will prompt the community to develop synthetic methods to solve the supply problem, which remain the number one hurdle for MNP drug development. In parallel, genomics-driven discovery will provide us with currently cryptic chemical diversity, and even assist with target and mechanism discovery. Synthetic biology methods will slowly but surely improve for the heterologous production of MNP, addressing the supply issue through biotechnological means. AI is expected to contribute and synergise with the field of MNP at all these levels, from metagenomics to structure and target identification, as well as production.
“Special” MNPs have intrinsic functions engineered by nature and evolution. They will continue to pave the way to new targets and be superior to unnatural approaches. As we coexist with nature, we learn more about the best approaches to deal with problems on this planet, including health solutions. We expect that marine sources will provide a rich supply of promising first-in-class therapeutic agents, assuming we can preserve the life-sustaining oceans and biological treasures contained in them, with new structural and biological characteristics123.
Key points
- Marine invertebrates and microorganisms produce novel marine natural products (MNPs) with potent and selective activity against many human diseases;
- Marine biodiversity is greater than that on land with more than 20 marine animal phyla having no terrestrial representatives;
- The greater the biological diversity investigated, the greater the chemical diversity that will be found — many of these marine-derived pharmaceuticals have novel mechanisms of action;
- About 15–20 compounds of marine origin have been approved for human use against cancer, pain, heart disease and viral infections, with many more in the pipeline;
- Aquaculture, chemical synthesis, and molecular genetics can supply material for clinical trials and clinical use;
- Antibody drug conjugate warheads developed from MNPs continue to feed the early clinical pipeline and offer exciting new avenues for MNP-based therapeutics;
- Although anti-cancer properties have been primarily studied, preclinical pharmacology of novel MNPs has demonstrated antibacterial, antifungal, antimalarial, antituberculosis, antiviral, antidiabetic, anti-inflammatory as well as immune and nervous system bioactivity;
- Understanding the mechanism of action added tremendous value to MNPs as they provide opportunities to decipher new biology that can be exploited to develop first-in-class therapeutics;
- Technical improvements have sped up the time taken to isolate and structurally characterise novel compounds whilst reducing effort wasted on the rediscovery of known compounds;
- These combined scientific advancements mean that pharmaceutical companies now regard marine natural product-derived pharmaceuticals with renewed interest.
Financial disclosure/conflict of interest statement/acknowledgements
The authors have no conflicts of interest except where explicitly stated.
MJ: This work was supported by a grant entitled ‘DEEPEND’ from the UK’s Department for Environment, Food and Rural Affairs Global Centre on Biodiversity for Climate.
CJ: This work was supported by grants PID2021-122732OB-C22 from MCIN/AEI/10.13039/ 501100011033/ FEDER “A way to make Europe” (AEI, Spanish State Agency for Research and FEDER Programme from the European Union). This publication is part of the work from COST Action CA18238 (Ocean4Biotech), funded by the European Cooperation in Science and Technology (COST) Program in the period 2019–2023. CJ was also supported by grants ED431C 2022/39 from Xunta de Galicia.
HL: Marine natural products drug discovery research by H.L. is supported by the National Institutes of Health (grants R01CA172310 and RM1GM145426) and the Debbie and Sylvia DeSantis Chair professorship. HL is co-founder of Oceanyx Pharmaceuticals, Inc. which licensed and optioned technologies and compounds based on marine natural products.
AMSM/MLP: This research was supported by Midwestern University Intramural Funds and the Midwestern University’s Office of Research.
DJN: Self-funded.
OTS: acknowledges support by EU funding within the MUR PNRR Extended Partnership initiative on Emerging Infectious Diseases (Project no. PE00000007, INF-ACT).
Author contributions
All authors contributed equally to the planning and writing of this article.
- 1.Newman DJ, Cragg GM. Natural Products as Sources of New Drugs over the Nearly Four Decades from 01/1981 to 09/2019. J Nat Prod. 2020;83(3):770-803. doi:10.1021/acs.jnatprod.9b01285
- 2.Marine Pharmacology. Marine Pharmacology. Accessed July 2024. https://www.marinepharmacology.org/
- 3.Blunt J, Buckingham J, Munro M. Taxonomy and Marine Natural Products Research. Handbook of Marine Natural Products. Published online 2012:3-54. doi:10.1007/978-90-481-3834-0_1
- 4.Briggs JC. Species Diversity: Land and Sea Compared. Systematic Biology. 1994;43(1):130. doi:10.2307/2413586
- 5.Ocean Biodiversity Information System. Ocean Biodiversity Information System. Accessed July 2024. www.obis.org
- 6.Williams DH, Stone MJ, Hauck PR, Rahman SK. Why Are Secondary Metabolites (Natural Products) Biosynthesized? J Nat Prod. 1989;52(6):1189-1208. doi:10.1021/np50066a001
- 7.Skropeta D. Deep-sea natural products. Nat Prod Rep. 2008;25(6):1131. doi:10.1039/b808743a
- 8.MarinLit. Royal Society of Chemistry. Accessed July 2024. https://marinlit.rsc.org/
- 9.Stone S, Newman DJ, Colletti SL, Tan DS. Cheminformatic analysis of natural product-based drugs and chemical probes. Nat Prod Rep. 2022;39(1):20-32. doi:10.1039/d1np00039j
- 10.Voser TM, Campbell MD, Carroll AR. How different are marine microbial natural products compared to their terrestrial counterparts? Nat Prod Rep. 2022;39(1):7-19. doi:10.1039/d1np00051a
- 11.Larsen AK, Galmarini CM, D’Incalci M. Unique features of trabectedin mechanism of action. Cancer Chemother Pharmacol. 2015;77(4):663-671. doi:10.1007/s00280-015-2918-1
- 12.Houssen WE, Jaspars M. Isolation of Marine Natural Products. Natural Products Isolation. Published online 2006:353-390. doi:10.1385/1-59259-955-9:353
- 13.Rodríguez J, Crews P, Jaspars M. Contemporary Strategies in Natural Products Structure Elucidation. Handbook of Marine Natural Products. Published online 2012:423-517. doi:10.1007/978-90-481-3834-0_7
- 14.Bergmann W, Feeney RJ. THE ISOLATION OF A NEW THYMINE PENTOSIDE FROM SPONGES1. J Am Chem Soc. 1950;72(6):2809-2810. doi:10.1021/ja01162a543
- 15.BERGMANN W, FEENEY RJ. CONTRIBUTIONS TO THE STUDY OF MARINE PRODUCTS. XXXII. THE NUCLEOSIDES OF SPONGES. I.1. J Org Chem. 1951;16(6):981-987. doi:10.1021/jo01146a023
- 16.BERGMANN W, BURKE DC. CONTRIBUTIONS TO THE STUDY OF MARINE PRODUCTS. XXXIX. THE NUCLEOSIDES OF SPONGES. III.1 SPONGOTHYMIDINE AND SPONGOURIDINE2. J Org Chem. 1955;20(11):1501-1507. doi:10.1021/jo01128a007
- 17.Hruba L, Das V, Hajduch M, Dzubak P. Nucleoside-based anticancer drugs: Mechanism of action and drug resistance. Biochemical Pharmacology. 2023;215:115741. doi:10.1016/j.bcp.2023.115741
- 18.Pizer LI, Cohen SS. Metabolism of Pyrimidine Arabinonucleosides and Cyclonucleosides in Escherichia coli. Journal of Biological Chemistry. 1960;235(8):2387-2392. doi:10.1016/s0021-9258(18)64632-0
- 19.Brown DM, Todd A, Varadarajan S. 462. Nucleotides. Part XXXVII. The structure of uridylic acids a and b, and a synthesis of spongouridine (3-β-<scp>D</scp>-arabofuranosyluracil). J Chem Soc. 1956;0(0):2388-2393. doi:10.1039/jr9560002388
- 20.Fox JJ, Yung N, Bendich A. Pyrimidine Nucleosides. II. The Synthesis of 1-β-D-Arabinofuranosylthymine (“Spongothymidine”)1. J Am Chem Soc. 1957;79(11):2775-2778. doi:10.1021/ja01568a030
- 21.Walwick E, Roberts W, Dekker C. Cyclisation during the phosphorylation of uridine and cytidine by polyphosphoric acid : a new route to the 02,2’-cyclonucleosides. Proc Chem Soc. 1959;84:415-423.
- 22.Lee WW, Benitez A, Goodman L, Baker BR. POTENTIAL ANTICANCER AGENTS.1 XL. SYNTHESIS OF THE β-ANOMER OF 9-(D-ARABINOFURANOSYL)-ADENINE. J Am Chem Soc. 1960;82(10):2648-2649. doi:10.1021/ja01495a070
- 23.Evans JS, Musser EA, Mengel GD, Forsblad KR, Hunter JH. Antitumor Activity of 1- -D-Arabinofuranosylcytosine Hydrochloride. Experimental Biology and Medicine. 1961;106(2):350-353. doi:10.3181/00379727-106-26335
- 24.Fermentation process for 9-(beta-d-arabinofuranosyl)adenine. Google Patents. Published 1967. Accessed July 2024. https://patents.google.com/patent/US3616208A/en
- 25.Cimino G, De Rosa S, De Stefano S. Antiviral agents from a gorgonian,Eunicella cavolini. Experientia. 1984;40(4):339-340. doi:10.1007/bf01952539
- 26.Bertin MJ, Schwartz SL, Lee J, et al. Spongosine Production by a Vibrio harveyi Strain Associated with the Sponge Tectitethya crypta. J Nat Prod. 2015;78(3):493-499. doi:10.1021/np5009762
- 27.Wilson MC, Mori T, Rückert C, et al. An environmental bacterial taxon with a large and distinct metabolic repertoire. Nature. 2014;506(7486):58-62. doi:10.1038/nature12959
- 28.Piel J. A polyketide synthase-peptide synthetase gene cluster from an uncultured bacterial symbiont of Paederus beetles. Proc Natl Acad Sci USA. 2002;99(22):14002-14007. doi:10.1073/pnas.222481399
- 29.Hemmerling F, Piel J. Strategies to access biosynthetic novelty in bacterial genomes for drug discovery. Nat Rev Drug Discov. 2022;21(5):359-378. doi:10.1038/s41573-022-00414-6
- 30.Rath CM, Janto B, Earl J, et al. Meta-omic Characterization of the Marine Invertebrate Microbial Consortium That Produces the Chemotherapeutic Natural Product ET-743. ACS Chem Biol. 2011;6(11):1244-1256. doi:10.1021/cb200244t
- 31.Schofield MM, Jain S, Porat D, Dick GJ, Sherman DH. Identification and analysis of the bacterial endosymbiont specialized for production of the chemotherapeutic natural product <scp>ET</scp>‐743. Environmental Microbiology. 2015;17(10):3964-3975. doi:10.1111/1462-2920.12908
- 32.Jiménez C. Marine Natural Products in Medicinal Chemistry. ACS Med Chem Lett. 2018;9(10):959-961. doi:10.1021/acsmedchemlett.8b00368
- 33.Dat TTH, Steinert G, Cuc NTK, Smidt H, Sipkema D. Bacteria Cultivated From Sponges and Bacteria Not Yet Cultivated From Sponges—A Review. Front Microbiol. 2021;12. doi:10.3389/fmicb.2021.737925
- 34.Chen J, Zhang P, Ye X, et al. The Structural Diversity of Marine Microbial Secondary Metabolites Based on Co-Culture Strategy: 2009–2019. Marine Drugs. 2020;18(9):449. doi:10.3390/md18090449
- 35.Schaufelberger DE, Koleck MP, Beutler JA, et al. The Large-Scale Isolation of Bryostatin 1 from Bugula neritina Following Current Good Manufacturing Practices. J Nat Prod. 1991;54(5):1265-1270. doi:10.1021/np50077a004
- 36.Tian Z, Lu XT, Jiang X, Tian J. Bryostatin-1: a promising compound for neurological disorders. Front Pharmacol. 2023;14. doi:10.3389/fphar.2023.1187411
- 37.Newman DJ. “From Large-Scale Collections to the Potential Use of Genomic Techniques for Supply of Drug Candidates.” Front Mar Sci. 2018;5. doi:10.3389/fmars.2018.00401
- 38.Hirata Y, Uemura D. Halichondrins – antitumor polyether macrolides from a marine sponge. Pure and Applied Chemistry. 1986;58(5):701-710. doi:10.1351/pac198658050701
- 39.Kawano S, Ito K, Yahata K, et al. A landmark in drug discovery based on complex natural product synthesis. Sci Rep. 2019;9(1). doi:10.1038/s41598-019-45001-9
- 40.Yu M, Littlefield B, Kishi Y. Discovery of E7389, a Fully Synthetic Macrocyclic Ketone Analog of Halichondrin B. Anticancer Agents from Natural Products. Published online June 13, 2005. doi:10.1201/9781420039658.ch13
- 41.Jacot W, Heudel P, Fraisse J, et al. Real‐life activity of eribulin mesylate among metastatic breast cancer patients in the multicenter national observational ESME program. Intl Journal of Cancer. 2019;145(12):3359-3369. doi:10.1002/ijc.32402
- 42.Erba E, Bergamaschi D, Bassano L, et al. Ecteinascidin-743 (ET-743), a natural marine compound, with a unique mechanism of action. European Journal of Cancer. 2001;37(1):97-105. doi:10.1016/s0959-8049(00)00357-9
- 43.ARAI T, TAKAHASHI K, ISHIGURO K, YAZAWA K. Increased production of saframycin A and isolation of saframycin S. J Antibiot. 1980;33(9):951-960. doi:10.7164/antibiotics.33.951
- 44.Cuevas C, Francesch A. Development of Yondelis® (trabectedin, ET-743). A semisynthetic process solves the supply problem. Nat Prod Rep. 2009;26(3):322. doi:10.1039/b808331m
- 45.Pettit GR, Kamano Y, Herald CL, et al. The isolation and structure of a remarkable marine animal antineoplastic constituent: dolastatin 10. J Am Chem Soc. 1987;109(22):6883-6885. doi:10.1021/ja00256a070
- 46.Pettit GR, Singh SB, Hogan F, et al. Antineoplastic agents. Part 189. The absolute configuration and synthesis of natural (-)-dolastatin 10. J Am Chem Soc. 1989;111(14):5463-5465. doi:10.1021/ja00196a061
- 47.Pettit G, Singh S, Inventors. Synthesis of Dolastatin 10. Google Patents. Published 1990. Accessed July 2024. https://patents.google.com/patent/US4978744A/en
- 48.Harrigan GG, Luesch H, Yoshida WY, et al. Symplostatin 1: A Dolastatin 10 Analogue from the Marine Cyanobacterium Symploca hydnoides. J Nat Prod. 1998;61(9):1075-1077. doi:10.1021/np980321c
- 49.Luesch H, Moore RE, Paul VJ, Mooberry SL, Corbett TH. Isolation of Dolastatin 10 from the Marine Cyanobacterium Symploca Species VP642 and Total Stereochemistry and Biological Evaluation of Its Analogue Symplostatin 1. J Nat Prod. 2001;64(7):907-910. doi:10.1021/np010049y
- 50.Engene N, Tronholm A, Salvador‐Reyes LA, Luesch H, Paul VJ. Caldora penicillata gen. nov., comb. nov. (Cyanobacteria), a pantropical marine species with biomedical relevance. Gabrielson P, ed. Journal of Phycology. 2015;51(4):670-681. doi:10.1111/jpy.12309
- 51.Salvador-Reyes LA, Engene N, Paul VJ, Luesch H. Targeted Natural Products Discovery from Marine Cyanobacteria Using Combined Phylogenetic and Mass Spectrometric Evaluation. J Nat Prod. 2015;78(3):486-492. doi:10.1021/np500931q
- 52.Kallifidas D, Dhakal D, Chen M, et al. Biosynthesis of Dolastatin 10 in Marine Cyanobacteria, a Prototype for Multiple Approved Cancer Drugs. Org Lett. 2024;26(7):1321-1325. doi:10.1021/acs.orglett.3c04083
- 53.Bennett G, Brown A, Mudd G, et al. MMAE Delivery Using the Bicycle Toxin Conjugate BT5528. Molecular Cancer Therapeutics. 2020;19(7):1385-1394. doi:10.1158/1535-7163.mct-19-1092
- 54.Shen L, Sun X, Chen Z, et al. ADCdb: the database of antibody–drug conjugates. Nucleic Acids Research. 2023;52(D1):D1097-D1109. doi:10.1093/nar/gkad831
- 55.FDA Approves Brentuximab Vedotin in Two Lymphoma Indications. The ASCO Post. Published 2011. Accessed July 2024. https://ascopost.com/issues/september-15-2011/fda-approves-brentuximab-vedotin-in-two-lymphoma-indications/#:~:text=The%20antibody%2Ddrug%20conjugate%20brentuximab,in%20more%20than%2030%20years
- 56.Brentuximab vedotin for treating CD30-positive Hodgkin lymphoma (CDF review of TA446). National Institute for Health and Care Excellence. Published 2017. Accessed July 2024. https://www.nice.org.uk/guidance/ta524/documents/final-appraisal-determination-document
- 57.FDA approves polatuzumab vedotin-piiq for diffuse large B-cell lymphoma. US FDA. Published 2019. Accessed July 2024. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-polatuzumab-vedotin-piiq-diffuse-large-b-cell-lymphoma
- 58.FDA grants accelerated approval to enfortumab vedotin-ejfv for metastatic urothelial cancer. US FDA. Published 2019. Accessed July 2024. https://www.fda.gov/drugs/resources-information-approved-drugs/fda-grants-accelerated-approval-enfortumab-vedotin-ejfv-metastatic-urothelial-cancer
- 59.Polatuzumab vedotin with rituximab and bendamustine for treating relapsed or refractory diffuse large B-cell lymphoma. National Institute for Health and Care Excellence. Published 2020. Accessed July 2024. https://www.nice.org.uk/guidance/ta649
- 60.Enfortumab vedotin for previously treated locally advanced or metastatic urothelial cancer (terminated appraisal). National Institute for Health and Care Excellence. Published 2022. Accessed July 2024. https://www.nice.org.uk/guidance/ta797
- 61.Tisotumab vedotin for treating recurrent or metastatic cervical cancer after systemic therapy. National Institute for Health and Care Excellence. Accessed July 2024. https://www.nice.org.uk/guidance/indevelopment/gid-ta10620
- 62.Drug Approval Package. Ziconotide Intrathecal Infusion. US FDA. Published 2004. Accessed July 2024. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2004/21-060_Prialt.cfm
- 63.Approved Drugs Lovaza. US FDA. Published 2004. Accessed July 2024. https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=overview.process&ApplNo=021654
- 64.Approved Drugs Vascepa. US FDA. Published 2012. Accessed July 2024. https://www.accessdata.fda.gov/scripts/cder/daf/index.cfm?event=overview.process&ApplNo=202057
- 65.Approved Drugs Epanova. US FDA. Published 2014. Accessed July 2024. https://www.accessdata.fda.gov/drugsatfda_docs/nda/2014/205060Orig1EpanovaTOC.cfm
- 66.EU/3/01/039 – orphan designation for treatment of soft tissue sarcoma. European Medicines Agency. Published 2007. Accessed July 2024. https://www.ema.europa.eu/en/medicines/human/orphan-designations/eu-3-01-039
- 67.Trabectedin for the treatment of advanced soft tissue sarcoma. National Institute for Health and Care Excellence. Published 2009. Accessed July 2024. https://www.nice.org.uk/guidance/ta185/documents/soft-tissue-sarcoma-trabectedin-final-appraisal-determination-fad2#:~:text=Trabectedin%20has%20a%20UK%20marketing,granted%20under%20’exceptional%20circumstances
- 68.Eribulin for the treatment of locally advanced or metastatic breast cancer. National Institute for Health and Care Excellence. Published 2011. Accessed July 2024. https://www.nice.org.uk/guidance/ta423/documents/final-appraisal-determination-document-2
- 69.Cheng X, Li J, Tanaka K, et al. MORAb-202, an Antibody–Drug Conjugate Utilizing Humanized Anti-human FRα Farletuzumab and the Microtubule-targeting Agent Eribulin, has Potent Antitumor Activity. Molecular Cancer Therapeutics. 2018;17(12):2665-2675. doi:10.1158/1535-7163.mct-17-1215
- 70.Australian Public Assessment Report for Plitidepsin. Australian Government. Published 2019. Accessed July 2024. https://www.tga.gov.au/sites/default/files/auspar-plitidepsin-190513.pdf
- 71.Aguareles J, Fernández PV, Carralón-González MM, et al. Outcomes and clinical characteristics of the compassionate use of plitidepsin for immunocompromised adult patients with COVID-19. International Journal of Infectious Diseases. 2023;135:12-17. doi:10.1016/j.ijid.2023.07.011
- 72.Hansen KØ, Hansen IKØ, Richard CS, Jenssen M, Andersen JH, Hansen EH. Antimicrobial Activity of Securamines From the Bryozoan Securiflustra securifrons. Natural Product Communications. 2021;16(2):1934578X2199618. doi:10.1177/1934578×21996180
- 73.Tripathi SK, Feng Q, Liu L, et al. Puupehenone, a Marine-Sponge-Derived Sesquiterpene Quinone, Potentiates the Antifungal Drug Caspofungin by Disrupting Hsp90 Activity and the Cell Wall Integrity Pathway. Mitchell AP, ed. mSphere. 2020;5(1). doi:10.1128/msphere.00818-19
- 74.Wright AE, Collins JE, Roberts B, et al. Antiplasmodial Compounds from Deep-Water Marine Invertebrates. Marine Drugs. 2021;19(4):179. doi:10.3390/md19040179
- 75.Liu Z, Wang Q, Li S, et al. Polypropionate Derivatives with Mycobacterium tuberculosis Protein Tyrosine Phosphatase B Inhibitory Activities from the Deep-Sea-Derived Fungus Aspergillus fischeri FS452. J Nat Prod. 2019;82(12):3440-3449. doi:10.1021/acs.jnatprod.9b00834
- 76.Lin SC, Lehman CW, Stewart AK, et al. Homoseongomycin, a compound isolated from marine actinomycete bacteria K3-1, is a potent inhibitor of encephalitic alphaviruses. Antiviral Research. 2021;191:105087. doi:10.1016/j.antiviral.2021.105087
- 77.Yang HW, Son M, Choi J, et al. Effect of Ishophloroglucin A, A Component of Ishige okamurae, on Glucose Homeostasis in the Pancreas and Muscle of High Fat Diet-Fed Mice. Marine Drugs. 2019;17(11):608. doi:10.3390/md17110608
- 78.Laborde RJ, Ishimura ME, Abreu-Butin L, et al. Sticholysins, pore-forming proteins from a marine anemone can induce maturation of dendritic cells through a TLR4 dependent-pathway. Molecular Immunology. 2021;131:144-154. doi:10.1016/j.molimm.2020.12.032
- 79.Daskalaki MG, Vyrla D, Harizani M, et al. Neorogioltriol and Related Diterpenes from the Red Alga Laurencia Inhibit Inflammatory Bowel Disease in Mice by Suppressing M1 and Promoting M2-Like Macrophage Responses. Marine Drugs. 2019;17(2):97. doi:10.3390/md17020097
- 80.Andrud K, Xing H, Gabrielsen B, et al. Investigation of the Possible Pharmacologically Active Forms of the Nicotinic Acetylcholine Receptor Agonist Anabaseine. Marine Drugs. 2019;17(11):614. doi:10.3390/md17110614
- 81.Rigby M, Bennett G, Chen L, et al. BT8009; A Nectin-4 Targeting Bicycle Toxin Conjugate for Treatment of Solid Tumors. Molecular Cancer Therapeutics. 2022;21(12):1747-1756. doi:10.1158/1535-7163.mct-21-0875
- 82.Domb C, Garcia JA, Barata PC, Mendiratta P, Rao S, Brown JR. Systematic review of recent advancements in antibody-drug and bicycle toxin conjugates for the treatment of urothelial cancer. Therapeutic Advances in Urology. 2024;16. doi:10.1177/17562872241249073
- 83.Gaudêncio SP, Bayram E, Lukić Bilela L, et al. Advanced Methods for Natural Products Discovery: Bioactivity Screening, Dereplication, Metabolomics Profiling, Genomic Sequencing, Databases and Informatic Tools, and Structure Elucidation. Marine Drugs. 2023;21(5):308. doi:10.3390/md21050308
- 84.Zhang G, Li J, Zhu T, Gu Q, Li D. Advanced tools in marine natural drug discovery. Current Opinion in Biotechnology. 2016;42:13-23. doi:10.1016/j.copbio.2016.02.021
- 85.Lebar MD, Heimbegner JL, Baker BJ. Cold-water marine natural products. Nat Prod Rep. 2007;24(4):774. doi:10.1039/b516240h
- 86.Skropeta D, Wei L. Recent advances in deep-sea natural products. Nat Prod Rep. 2014;31(8):999-1025. doi:10.1039/c3np70118b
- 87.Epstein S. The phenomenon of microbial uncultivability. Current Opinion in Microbiology. 2013;16(5):636-642. doi:10.1016/j.mib.2013.08.003
- 88.Zhang ZF, Liu F, Liu LR, et al. Culturing the uncultured marine fungi in the omics age: Opportunities and challenges. Fungal Biology Reviews. 2024;48:100353. doi:10.1016/j.fbr.2023.100353
- 89.Nichols D, Cahoon N, Trakhtenberg EM, et al. Use of Ichip for High-Throughput In Situ Cultivation of “Uncultivable” Microbial Species. Appl Environ Microbiol. 2010;76(8):2445-2450. doi:10.1128/aem.01754-09
- 90.Liu X, Wang M, Nie Y, Wu XL. Isolation Chip Increases Culturable Bacterial Diversity and Reduces Cultivation Bias. Curr Microbiol. 2021;78(5):2025-2032. doi:10.1007/s00284-021-02474-0
- 91.Riguera R. Isolating bioactive compounds from marine organisms. J Mar Biotechnol. 1997;76:2445-2450. https://www.researchgate.net/profile/Ricardo-Riguera/publication/226373771_Isolating_bioactive_compounds_from_marine_organisms/links/5640604408ae34e98c4e7d5e/Isolating-bioactive-compounds-from-marine-organisms.pdf?_tp=eyJjb250ZXh0Ijp7ImZpcnN0UGFnZSI6InB1YmxpY2F0aW9uIiwicGFnZSI6InB1YmxpY2F0aW9uIn19
- 92.Gaudêncio SP, Pereira F. Dereplication: racing to speed up the natural products discovery process. Nat Prod Rep. 2015;32(6):779-810. doi:10.1039/c4np00134f
- 93.Watrous J, Roach P, Alexandrov T, et al. Mass spectral molecular networking of living microbial colonies. Proc Natl Acad Sci USA. 2012;109(26). doi:10.1073/pnas.1203689109
- 94.Wang M, Carver JJ, Phelan VV, et al. Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking. Nat Biotechnol. 2016;34(8):828-837. doi:10.1038/nbt.3597
- 95.Nothias LF, Petras D, Schmid R, et al. Feature-based molecular networking in the GNPS analysis environment. Nat Methods. 2020;17(9):905-908. doi:10.1038/s41592-020-0933-6
- 96.Bruguière A, Derbré S, Bréard D, Tomi F, Nuzillard JM, Richomme P. 13C NMR Dereplication Using MixONat Software: A Practical Guide to Decipher Natural Products Mixtures. Planta Med. 2021;87(12/13):1061-1068. doi:10.1055/a-1470-0446
- 97.Paytubi S, de La Cruz M, Tormo JR, et al. A High-Throughput Screening Platform of Microbial Natural Products for the Discovery of Molecules with Antibiofilm Properties against Salmonella. Front Microbiol. 2017;8. doi:10.3389/fmicb.2017.00326
- 98.Molinski TF. NMR of natural products at the ‘nanomole-scale.’ Nat Prod Rep. 2010;27(3):321. doi:10.1039/b920545b
- 99.Hubert J, Nuzillard JM, Renault JH. Dereplication strategies in natural product research: How many tools and methodologies behind the same concept? Phytochem Rev. 2015;16(1):55-95. doi:10.1007/s11101-015-9448-7
- 100.Medema MH, Kottmann R, Yilmaz P, et al. Minimum Information about a Biosynthetic Gene cluster. Nat Chem Biol. 2015;11(9):625-631. doi:10.1038/nchembio.1890
- 101.Navarro-Muñoz JC, Selem-Mojica N, Mullowney MW, et al. A computational framework to explore large-scale biosynthetic diversity. Nat Chem Biol. 2019;16(1):60-68. doi:10.1038/s41589-019-0400-9
- 102.Albarano L, Esposito R, Ruocco N, Costantini M. Genome Mining as New Challenge in Natural Products Discovery. Marine Drugs. 2020;18(4):199. doi:10.3390/md18040199
- 103.Medema MH, Blin K, Cimermancic P, et al. antiSMASH: rapid identification, annotation and analysis of secondary metabolite biosynthesis gene clusters in bacterial and fungal genome sequences. Nucleic Acids Research. 2011;39(suppl_2):W339-W346. doi:10.1093/nar/gkr466
- 104.Oulas A, Pavloudi C, Polymenakou P, et al. Metagenomics: Tools and Insights for Analyzing Next-Generation Sequencing Data Derived from Biodiversity Studies. Bioinform Biol Insights. 2015;9:BBI.S12462. doi:10.4137/bbi.s12462
- 105.Montalbán-López M, Scott TA, Ramesh S, et al. New developments in RiPP discovery, enzymology and engineering. Nat Prod Rep. 2021;38(1):130-239. doi:10.1039/d0np00027b
- 106.Trosset JY, Carbonell P. Synthetic biology for pharmaceutical drug discovery. DDDT. Published online December 2015:6285. doi:10.2147/dddt.s58049
- 107.Prihoda D, Maritz JM, Klempir O, et al. The application potential of machine learning and genomics for understanding natural product diversity, chemistry, and therapeutic translatability. Nat Prod Rep. 2021;38(6):1100-1108. doi:10.1039/d0np00055h
- 108.Walker AS, Clardy J. A Machine Learning Bioinformatics Method to Predict Biological Activity from Biosynthetic Gene Clusters. J Chem Inf Model. 2021;61(6):2560-2571. doi:10.1021/acs.jcim.0c01304
- 109.Hu G, Qiu M. Machine learning-assisted structure annotation of natural products based on MS and NMR data. Nat Prod Rep. 2023;40(11):1735-1753. doi:10.1039/d3np00025g
- 110.SMART 2.1 Drag and Drop. UC San Diego. Accessed July 2024. http://smart.ucsd.edu/classic
- 111.Kim HW, Zhang C, Cottrell GW, Gerwick WH. SMART‐Miner: A convolutional neural network‐based metabolite identification from 1H‐13C HSQC spectra. Magnetic Reson in Chemistry. 2021;60(11):1070-1075. doi:10.1002/mrc.5240
- 112.Complex Mixture Analysis by NMR. Ohio State University. Accessed July 2024. https://spin.ccic.osu.edu/index.php/colmar
- 113.Leao TF, Clark CM, Bauermeister A, et al. Quick-start infrastructure for untargeted metabolomics analysis in GNPS. Nat Metab. 2021;3(7):880-882. doi:10.1038/s42255-021-00429-0
- 114.Mullowney MW, Duncan KR, Elsayed SS, et al. Artificial intelligence for natural product drug discovery. Nat Rev Drug Discov. 2023;22(11):895-916. doi:10.1038/s41573-023-00774-7
- 115.Jumper J, Evans R, Pritzel A, et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583-589. doi:10.1038/s41586-021-03819-2
- 116.Kleigrewe K, Almaliti J, Tian IY, et al. Combining Mass Spectrometric Metabolic Profiling with Genomic Analysis: A Powerful Approach for Discovering Natural Products from Cyanobacteria. J Nat Prod. 2015;78(7):1671-1682. doi:10.1021/acs.jnatprod.5b00301
- 117.Gerwick WH. The Face of a Molecule. J Nat Prod. 2017;80(9):2583-2588. doi:10.1021/acs.jnatprod.7b00624
- 118.Luesch H, MacMillan JB. Targeting and extending the eukaryotic druggable genome with natural products. Nat Prod Rep. 2020;37(6):744-746. doi:10.1039/d0np90021d
- 119.Shen L, Pelletier J. Selective targeting of the DEAD-box RNA helicase eukaryotic initiation factor (eIF) 4A by natural products. Nat Prod Rep. 2020;37(5):609-616. doi:10.1039/c9np00052f
- 120.Childs-Disney JL, Yang X, Gibaut QMR, Tong Y, Batey RT, Disney MD. Targeting RNA structures with small molecules. Nat Rev Drug Discov. 2022;21(10):736-762. doi:10.1038/s41573-022-00521-4
- 121.Potts MB, McMillan EA, Rosales TI, et al. Mode of action and pharmacogenomic biomarkers for exceptional responders to didemnin B. Nat Chem Biol. 2015;11(6):401-408. doi:10.1038/nchembio.1797
- 122.Liang X, Luo D, Luesch H. Advances in exploring the therapeutic potential of marine natural products. Pharmacological Research. 2019;147:104373. doi:10.1016/j.phrs.2019.104373
- 123.Rogers AD, Baco A, Escobar-Briones E, et al. Marine Genetic Resources in Areas Beyond National Jurisdiction: Promoting Marine Scientific Research and Enabling Equitable Benefit Sharing. Front Mar Sci. 2021;8. doi:10.3389/fmars.2021.667274