Victor Habbick Visions / Science Photo Library
In the UK, the proportion of older adults in the population is rising steadily. It is projected that, by 2050, one in four people will be aged 65 years and over, compared with one in five in 2018[1]. And, with an ageing population comes an increase in the prevalence of multimorbidities and polypharmacy[2].
Polypharmacy, commonly defined as the regular use of five or more medicines concomitantly, is associated with an increased likelihood of significant negative health outcomes[3–5]. Older people are at increased risk of adverse drug reactions (ADRs) and one in five hospital admissions in patients aged over 65 years is attributed to ADRs[6].
Risk factors for ADRs — such as age, renal function and comorbidities — are often considered by prescribers in medication reviews, but much more can be done to reduce the risk of harm and improve the efficacy of medicines. In particular, there is a growing body of evidence to support the use of pharmacogenomic testing to predict an individual’s response to many commonly used medicines and this could be an extremely useful tool when it comes to optimising treatment and reducing the risk of ADRs in older people[7].
Actionable variants
There have been rapid advances in the development of genotyping technologies that can quickly identify individual variations in key genes that affect a person’s ability to metabolise, or respond to, certain medicines[8].
It is estimated that more than 90% of individuals carry at least one genetic variant that is ‘actionable’ and could help inform a prescribing decision
It is estimated that more than 90% of individuals carry at least one genetic variant that is ‘actionable’ and could help inform a prescribing decision[9–11]. The results from pharmacogenomic tests could therefore enable prescribers to ensure that the right medicine is given at the right dose to the right patient.
The NHS is committed to embedding pharmacogenomic testing into routine practice by 2025, and the available evidence suggests that incorporating pharmacogenomic testing into the care of older people specifically will reduce ADRs and unplanned hospital admissions[12,13].
Our research
Comprising pharmacists and academics from the Medicines Optimisation Research Group at the University of Bradford, in collaboration with Leeds Teaching Hospitals NHS Trust (LTHT) and the University of Leeds, our research group is actively investigating how pharmacogenomic testing could be put into practice in the NHS.
The group is currently completing a retrospective cross-sectional study of 59,973 hospital admissions of people aged 65 years and over to LTHT throughout 2018 and 2019 and the 560,163 medicines that were prescribed to them.
As expected, polypharmacy was high, with 83% of patients taking five or more medicines and 43% taking ten or more medicines. Our analysis also showed that over one in five (22%) medicines being taken on admission had known pharmacogenomic associations, with 63 different medicines with known pharmacogenomic associations being used.
Many of the medicines identified as having pharmacogenomic links are also considered ‘high risk’ regarding their potential to contribute to unplanned hospital admissions
On average, each patient was taking nine medicines, two of which were actionable from a pharmacogenomics perspective.
Many of the medicines identified as having pharmacogenomic links are also considered ‘high risk’ regarding their potential to contribute to unplanned hospital admissions[14]. These include antiplatelets, anticoagulants, analgesics and antidepressants (see Table 1[15]). The Clinical Pharmacogenetics Implementation Consortium (CPIC) evaluates the level of existing clinical and research evidence for drug–gene pairs; those at level A or level B have potential prescribing recommendations[15].
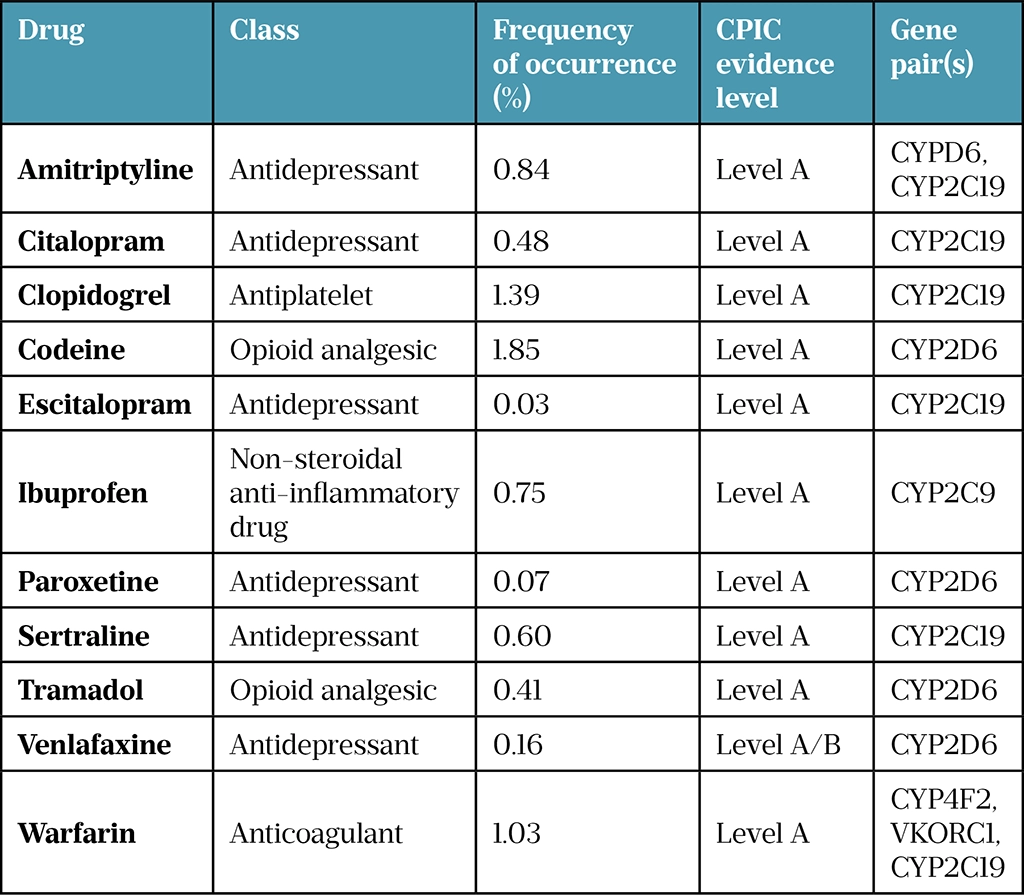
In the following sections, we will look at three of the most prevalent medicine groups in our sample and assess the evidence for intervention based on pharmacogenomic testing.
Antiplatelets and anticoagulants
Prescriptions for clopidogrel and warfarin accounted for between 1.0% and 1.4% (5,746–7,767) of all prescriptions for patients aged 65 years and over who were admitted to LTHT. Both of these medicines have actionable pharmacogenomic associations with strong CPIC recommendations to change prescribing[16,17].
For patients initiated on clopidogrel, a change in medication — to ticagrelor or prasugrel, for example — is recommended for poor and intermediate metabolisers of clopidogrel to improve the efficacy of antiplatelet therapy[18,19]. Point-of-care tests to identify the genetic variants that affect clopidogrel metabolism have already been developed and have a 55-minute turnaround time, meaning that results can be acted upon during inpatient admission and outpatient review, potentially in primary care and community pharmacy settings[20–26].
Like blood glucose monitoring, point-of-care testing can also be carried out by healthcare professionals without formal laboratory training
Like blood glucose monitoring, point-of care testing can also be carried out by healthcare professionals without formal laboratory training. Implementing pharmacogenomic testing for the 4–30% of patients initiated on clopidogrel who do not derive therapeutic benefit owing to the loss-of-function gene variant they carry, could be easily incorporated into patient care pathways, although the practical application, cost and clinical utility would need to be assessed first[27].
For warfarin, dose adjustment rather than medication switching is recommended for patients with certain genetic variants that result in ADRs or poor drug efficacy[16]. However, recently there has been a rapid decline in warfarin initiation owing to a move towards direct oral anticoagulants, which may reduce the need for routine pharmacogenomic testing in this cohort[28]. It continues to be used for some indications, such as for patients with mechanical heart valves; in these cases, prospective pharmacogenomic testing may be helpful to define the dose range.
High-risk analgesics
Pain management in older people can be complicated by age-related ADRs as well as polypharmacy and multimorbidity[3]. Between 0.4% to 1.9% (2,294 to 10,369) of all prescriptions in the study sample contained codeine, tramadol or ibuprofen. These have pharmacogenomic associations and are identified as ‘high risk’ for causing medicine-related hospital admissions.
Codeine and tramadol are opioid analgesics used to treat mild-to-moderate pain and moderate-to-severe pain, respectively. Both are metabolised to their active metabolites by cytochrome P450 2D6 (CYP2D6) enzymes, encoded by the highly polymorphic CYP2D6 gene. Globally, up to 7% of individuals are poor metabolisers of codeine and tramadol, leading to reduced efficacy, while 2% are ultra-rapid metabolisers, leading to an increased risk of opiate toxicity[8,29–31]. As a result, there are strong recommendations to avoid codeine/tramadol use in ultra-rapid and poor metabolisers to help avoid these outcomes[8].
There is evidence that pharmacogenomic information could be used to alter opioid prescribing behaviours and as an additional tool to optimise medicines while considering other risk factors, such as drug–drug interactions that also contribute to drug response[32].
After aspirin and diclofenac, ibuprofen is the third most common non-steroidal anti-inflammatory drug (NSAID) to cause medicine-related hospital admission owing to its gastrointestinal effects, such as bleeding and peptic ulceration, as well as renal impairment[14]. Up to 4% of individuals globally are CYP2C9 poor metabolisers, resulting in higher plasma concentrations of ibuprofen and an increased risk of severe toxicity[29,33]. However, there is currently no drug labelling to highlight this pharmacogenomic association and only moderate recommendations for dose adjustments, primarily owing to the lack of consensus on the clinical evidence supporting pharmacogenomic-guided prescribing recommendations[34]. Concomitant use of ibuprofen with other CYP2C9 substrates, such as warfarin, can increase the risk for ADRs, such as bleeding.
High-risk antidepressants
Just under 1% (4,698) of all prescriptions in the study sample were for antidepressants amitriptyline, venlafaxine, paroxetine, sertraline, citalopram and escitalopram. Patients are susceptible to antidepressant ADRs owing to the trial-and-error approach to prescribing for depression[35,36]. However, this could be mitigated by pharmacogenomic testing to predict a patient’s response to medicines. All of these antidepressants interact with CYP2D6 or CYP2C19 gene variants. Between 1–15% and 1–20% of patients are poor metabolisers and ultra-rapid metabolisers of tricyclic antidepressants, respectively[37]. For selective serotonin reuptake inhibitors (SSRIs), about 2–15% and 5–30% of patients are poor metabolisers and ultra-rapid metabolisers, respectively[38]. Unlike SSRIs and tricyclic antidepressants, venlafaxine does not have pharmacogenomic prescribing guidelines.
Between 1–15% and 1–20% of patients are poor metabolisers and ultra-rapid metabolisers of tricyclic antidepressants, respectively
Amitriptyline is a tricyclic antidepressant and the most common antidepressant in the study sample. It is used as an alternative medicine for severe chronic depressive symptoms when patients have been unresponsive to SSRIs[39]. Amitriptyline has numerous ADRs, such as anticholinergic syndrome and drowsiness that can lead to falls, with older people at an increased risk owing to increased prevalence of polypharmacy, frailty and cognitive decline[40]. In this drug–gene interaction, variants of the CYP2D6 gene affect the clearance of amitriptyline. As a result, there are strong recommendations to avoid its use in CYP2D6 poor metabolisers owing to the increased risk of ADRs and to consider alternative medication in CYP2D6 ultra-rapid metabolisers owing to reduced drug efficacy and the potential for therapeutic failure[37]. Amitriptyline is mostly used for neuropathic pain, and pharmacogenomic-based dose changes are not currently recommended for this indication since the drug is used at a lower dosage, meaning patients are less likely to experience ADRs owing to poor drug clearance. There is also insufficient data describing the use of amitriptyline for neuropathic pain in patients that are CYP2D6 ultra-rapid metabolisers[37].
SSRIs present in the dataset included paroxetine, sertraline, citalopram and escitalopram. Apart from escitalopram, these SSRIs can be used in first episode depression[39]. Paroxetine is extensively metabolised by CYP2D6 enzymes and ultra-rapid metabolisers will have reduced plasma concentrations of the active compound compared with more efficient metabolisers, increasing the risk of treatment failure[38]. However, since there are insufficient data to recommend an initial paroxetine dose for CYP2D6 ultra-rapid metabolisers, there is a strong recommendation to avoid it altogether in these patients[38]. The same bypass approach is recommended in CYP2C19 ultra-rapid metabolisers of citalopram and escitalopram where an alternative SSRI, not extensively metabolised by CYP2C19, is recommended[38]. Sertraline is also metabolised by CYP2C19 but differences in opinion exist in prescribing recommendations for poor and ultra-rapid metabolisers[38].
The future
In conclusion, patients are commonly taking high-risk medicines with pharmacogenomic associations when they are admitted to hospital. Results from our retrospective cross-sectional study, alongside the published evidence, suggest that carrying out point-of-care pharmacogenomic testing prior to initiating these medicines in older people could effectively optimise medicines and reduce ADRs. However, there are numerous challenges for effective implementation of this type of testing.
Carrying out point-of-care pharmacogenomic testing prior to initiating these medicines in older people could effectively optimise medicines and reduce ADRs
A joint report from the Royal College of Physicians and British Pharmacological Society, published in March 2022, highlighted key areas that require consideration, including standardising the consent process; storing and returning pharmacogenomic results in a user-friendly manner; building patient trust; and increasing the confidence of healthcare prescribers in pharmacogenomics[7].
Furthermore, current approaches to point-of-care testing could be restrictive in the genetic variants they test for as the frequency of genetic variants can differ significantly between ethnicities. A panel test approach, whereby multiple variants for multiple medicines are tested together, could provide the coverage required for testing variants that interact with the medicines discussed in this article.
Pharmacy teams
The expertise of pharmacists will be invaluable in delivering pharmacogenomic-guided medicines optimisation, for instance, when conducting structured medication reviews in England and under independent prescribing initiatives in Scotland and Wales. However, major initiatives in training and education are required to enable healthcare professionals to support improvements in personalised patient care led by genetic information[41]. This must happen as a matter of urgency, so that the health service can make effective use of pharmacogenomic testing to improve the care of older people.
Acknowledgement: This research was supported by the National Institute for Health Research Yorkshire and Humber Patient Safety Translational Research Centre. Gurdeep S. Sagoo was supported by the National Institute for Health Research Leeds In vitro Diagnostics Co-operative. This article presents independent research funded by Leeds Teaching Hospitals NHS Trust and the University of Bradford.
- 1Overview of the UK population: August 2019. Office of National Statistics. 2019.https://www.ons.gov.uk/peoplepopulationandcommunity/populationandmigration/populationestimates/articles/overviewoftheukpopulation/august2019#the-uks-population-is-ageing (accessed 5 Jul 2022).
- 2Moody A, Mindell J, Faulding S. Health Survey for England 2016 Prescribed medicines. NHS Digital. 2017.http://healthsurvey.hscic.gov.uk/media/63790/HSE2016-pres-med.pdf (accessed 5 Jul 2022).
- 3Masnoon N, Shakib S, Kalisch-Ellett L, et al. What is polypharmacy? A systematic review of definitions. BMC Geriatr. 2017;17. doi:10.1186/s12877-017-0621-2
- 4Hilmer S, Gnjidic D. The Effects of Polypharmacy in Older Adults. Clin Pharmacol Ther. 2008;85:86–8. doi:10.1038/clpt.2008.224
- 5Maher RL, Hanlon J, Hajjar ER. Clinical consequences of polypharmacy in elderly. Expert Opinion on Drug Safety. 2013;13:57–65. doi:10.1517/14740338.2013.827660
- 6Government pledges to reduce overprescribing of medicines. GOV.UK. 2021.https://www.gov.uk/government/news/government-pledges-to-reduce-overprescribing-of-medicines#:~:text=Ministers%20are%20announcing%20action%20to,Chief%20Pharmaceutical%20Officer%20for%20England. (accessed 5 Jul 2022).
- 7Personalised prescribing: using pharmacogenomics to improve patient outcomes. Royal College of Physicians. 2022.https://www.rcp.ac.uk/projects/outputs/personalised-prescribing-using-pharmacogenomics-improve-patient-outcomes (accessed 5 Jul 2022).
- 8Crews KR, Monte AA, Huddart R, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2D6 , OPRM1 , and COMT Genotypes and Select Opioid Therapy. Clin. Pharmacol. Ther. 2021;110:888–96. doi:10.1002/cpt.2149
- 9Van Driest S, Shi Y, Bowton E, et al. Clinically Actionable Genotypes Among 10,000 Patients With Preemptive Pharmacogenomic Testing. Clin Pharmacol Ther. 2013;95:423–31. doi:10.1038/clpt.2013.229
- 10Ji Y, Skierka JM, Blommel JH, et al. Preemptive Pharmacogenomic Testing for Precision Medicine. The Journal of Molecular Diagnostics. 2016;18:438–45. doi:10.1016/j.jmoldx.2016.01.003
- 11Chanfreau-Coffinier C, Hull LE, Lynch JA, et al. Projected Prevalence of Actionable Pharmacogenetic Variants and Level A Drugs Prescribed Among US Veterans Health Administration Pharmacy Users. JAMA Netw Open. 2019;2:e195345. doi:10.1001/jamanetworkopen.2019.5345
- 12David V, Fylan B, Bryant E, et al. An Analysis of Pharmacogenomic-Guided Pathways and Their Effect on Medication Changes and Hospital Admissions: A Systematic Review and Meta-Analysis. Front. Genet. 2021;12. doi:10.3389/fgene.2021.698148
- 13Youssef E, Kirkdale CL, Wright DJ, et al. Estimating the potential impact of implementing pre‐emptive pharmacogenetic testing in primary care across the UK. Br J Clin Pharmacol. 2021;87:2907–25. doi:10.1111/bcp.14704
- 14Pirmohamed M, James S, Meakin S, et al. Adverse drug reactions as cause of admission to hospital: prospective analysis of 18 820 patients. BMJ. 2004;329:15–9. doi:10.1136/bmj.329.7456.15
- 15Caudle KE, Gammal RS, Whirl-Carrillo M, et al. Evidence and resources to implement pharmacogenetic knowledge for precision medicine. American Journal of Health-System Pharmacy. 2016;73:1977–85. doi:10.2146/ajhp150977
- 16Johnson J, Caudle K, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline for Pharmacogenetics-Guided Warfarin Dosing: 2017 Update. Clin. Pharmacol. Ther. 2017;102:397–404. doi:10.1002/cpt.668
- 17Lee CR, Luzum JA, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium Guideline for CYP2C19 Genotype and Clopidogrel Therapy: 2022 Update. Clin Pharma and Therapeutics. 2022. doi:10.1002/cpt.2526
- 18Klein MD, Williams AK, Lee CR, et al. Clinical Utility of CYP2C19 Genotyping to Guide Antiplatelet Therapy in Patients With an Acute Coronary Syndrome or Undergoing Percutaneous Coronary Intervention. ATVB. 2019;39:647–52. doi:10.1161/atvbaha.118.311963
- 19Peyracchia M, Saglietto A, Biolè C, et al. Efficacy and Safety of Clopidogrel, Prasugrel and Ticagrelor in ACS Patients Treated with PCI: A Propensity Score Analysis of the RENAMI and BleeMACS Registries. Am J Cardiovasc Drugs. 2019;20:259–69. doi:10.1007/s40256-019-00373-1
- 20Spartan RX point-of-care CYP2C19 test to guide treatment in acute coronary syndrome. National Institute for Health and Care Excellence. 2020.https://www.nice.org.uk/advice/mib223 (accessed 5 Jul 2022).
- 21Claassens DMF, Vos GJA, Bergmeijer TO, et al. A Genotype-Guided Strategy for Oral P2Y12 Inhibitors in Primary PCI. N Engl J Med. 2019;381:1621–31. doi:10.1056/nejmoa1907096
- 22Bergmeijer TO, Vos GJ, Claassens DM, et al. Feasibility and implementation of CYP2C19 genotyping in patients using antiplatelet therapy. Pharmacogenomics. 2018;19:621–8. doi:10.2217/pgs-2018-0013
- 23Cavallari LH, Franchi F, Rollini F, et al. Clinical implementation of rapid CYP2C19 genotyping to guide antiplatelet therapy after percutaneous coronary intervention. J Transl Med. 2018;16. doi:10.1186/s12967-018-1469-8
- 24Zhou Y, Armstead A, Coshatt G, et al. Comparison of Two Point-of-Care CYP2C19 Genotyping Assays for Genotype-Guided Antiplatelet Therapy. Ann Clin Lab Sci 2017;47:738–43.https://www.ncbi.nlm.nih.gov/pubmed/29263048
- 25Koltowski L, Tomaniak M, Aradi D, et al. Optimal aNtiplatelet pharmacotherapy guided by bedSIDE genetic or functional TESTing in elective PCI patients: A pilot study: ONSIDE TEST pilot. Cardiol J 2017;24:284–92. doi:10.5603/CJ.a2017.0026
- 26Choi J, Kim B, Woo K, et al. The Diagnostic Utility of the Point-of-Care CYP2C19 Genotyping Assay in Patients with Acute Coronary Syndrome Dosing Clopidogrel: Comparison with Platelet Function Test and SNP Genotyping. Ann Clin Lab Sci 2016;46:489–94.https://www.ncbi.nlm.nih.gov/pubmed/27650615
- 27Nguyen TA, Diodati JG, Pharand C. Resistance to clopidogrel: A review of the evidence. Journal of the American College of Cardiology. 2005;45:1157–64. doi:10.1016/j.jacc.2005.01.034
- 28Loo SY, Dell’Aniello S, Huiart L, et al. Trends in the prescription of novel oral anticoagulants in UK primary care. Br J Clin Pharmacol. 2017;83:2096–106. doi:10.1111/bcp.13299
- 29Gene-specific Information Tables for CYP2D6. PHARMGKB. https://www.pharmgkb.org/page/cyp2d6RefMaterials (accessed 5 Jul 2022).
- 30Dean L, Kane M, Pratt V, et al. gtrbook. Published Online First: 30 March 2021.http://www.ncbi.nlm.nih.gov/books/NBK100662/
- 31Dean L, Kane M, Pratt V, et al. gtrbook. Published Online First: 20 July 2021.http://www.ncbi.nlm.nih.gov/books/NBK315950/
- 32Fulton CR, Zang Y, Desta Z, et al. Drug–gene and drug–drug interactions associated with tramadol and codeine therapy in the INGENIOUS trial. Pharmacogenomics. 2019;20:397–408. doi:10.2217/pgs-2018-0205
- 33Theken KN, Lee CR, Gong L, et al. Clinical Pharmacogenetics Implementation Consortium Guideline (CPIC) for CYP2C9 and Nonsteroidal Anti‐Inflammatory Drugs. Clin. Pharmacol. Ther. 2020;108:191–200. doi:10.1002/cpt.1830
- 34Abdullah-Koolmees H, van Keulen AM, Nijenhuis M, et al. Pharmacogenetics Guidelines: Overview and Comparison of the DPWG, CPIC, CPNDS, and RNPGx Guidelines. Front. Pharmacol. 2021;11. doi:10.3389/fphar.2020.595219
- 35Lombard J, Doraiswamy PM. What is the role of pharmacogenetics in clinical psychiatry? Expert Opinion on Drug Metabolism & Toxicology. 2012;9:1–4. doi:10.1517/17425255.2013.733696
- 36Virelli CR, Mohiuddin AG, Kennedy JL. Barriers to clinical adoption of pharmacogenomic testing in psychiatry: a critical analysis. Transl Psychiatry. 2021;11. doi:10.1038/s41398-021-01600-7
- 37Hicks J, Sangkuhl K, Swen J, et al. Clinical pharmacogenetics implementation consortium guideline (CPIC) for CYP2D6 and CYP2C19 genotypes and dosing of tricyclic antidepressants: 2016 update. Clin. Pharmacol. Ther. 2017;102:37–44. doi:10.1002/cpt.597
- 38Hicks J, Bishop J, Sangkuhl K, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) Guideline forCYP2D6andCYP2C19Genotypes and Dosing of Selective Serotonin Reuptake Inhibitors. Clin. Pharmacol. Ther. 2015;98:127–34. doi:10.1002/cpt.147
- 39Depression in adults: treatment and management. National Institute for Health and Care Excellence. 2022.https://www.nice.org.uk/guidance/ng222 (accessed 5 Jul 2022).
- 40Amitriptyline hydrochloride: side effects. British National Formulary. https://bnf.nice.org.uk/drugs/amitriptyline-hydrochloride/#side-effects (accessed 5 Jul 2022).
- 41Owusu-Obeng A, Weitzel KW, Hatton RC, et al. Emerging Roles for Pharmacists in Clinical Implementation of Pharmacogenomics. Pharmacotherapy. 2014;34:1102–12. doi:10.1002/phar.1481