Abstract
Growing evidence has highlighted the potentially significant impact of drug–microbiome interactions on patient care. It is possible that hundreds of drugs alter the composition of the microbiome, including many drugs with non-microbial targets. Drug-induced alteration of the microbiome could increase patients’ risk of dysbiosis, a state of microbiome unbalance that increases the chance of disease. Further, a drug’s pharmacokinetics and pharmacodynamics can be altered by the microbiome via direct (e.g. biotransformation or bioaccumulation) and indirect processes. Although these interactions are potentially important for patient health and therapeutic success, they are rarely considered during drug development or in clinical practice. Healthcare professionals working across sectors should consider drug–microbiome interactions to improve patient outcomes. This review provides an overview of the current evidence relating to drug–microbiome interactions, and describes how healthcare professionals working in clinical settings, academia, policy and drug development can immediately begin to address drug–microbiome interactions as an integral part of their roles.
Key words: biopharmaceutics; biotechnology; drug discovery and development; drug–microbiome relationship; future pharmacist roles; microbiota; pharmaceutical industry.
Key points
- The human microbiome is composed of trillions of microorganisms, including bacteria, fungi, archaea and viruses. Gut bacteria alone may encode up to 150 times more genes than their human hosts;
- Drugs can affect microbiome composition, including those with intended and unintended antimicrobial actions. These effects can form part of drugs’ therapeutic activity or may promote disease;
- The broad metabolic capacity of the gut microbiome can alter drugs’ pharmacokinetics directly and indirectly. More than 150 drugs are now known to be susceptible to chemical transformation or accumulation by intestinal bacteria;
- Healthcare professionals working in clinical settings may be increasingly required to consider drug–microbiome interactions; for example, when answering patients’ questions or managing the prescription of medicines;
- In the future, new technology may enable professionals to personalise patients’ dosing regimens based on their predicted drug–microbiome interactions;
- Professionals working in drug development should consider screening for drug–microbiome interactions at pre-clinical phases; this may identify potential challenges or opportunities at an early stage.
Introduction
Emerging evidence revealing important interactions between the microbiome, drugs and human health could significantly alter how medicines are developed and prescribed in the future. It is now recognised that drugs and the microbiome can have bidirectional relationships: drugs may affect microbiome composition, possibly leading to changes in host health, and the microbiome can correspondingly affect drugs, possibly promoting pharmacokinetic (PK) and/or pharmacodynamic (PD) variability between and within individuals[1]. As early as 2010, the role of the microbiome in modulating PK and PD has been defined as pharmacomicrobiomics, which represents a growing field of research working to characterise and even predict drug-microbiome interactions[2,3]. Similarly, the fields of pharmacometabolomics and pharmacometabonomics describe the measurement and analysis, respectively, of how metabolites (in this case, microbial metabolites) affect drug response[4]. Microbiota activity across the human body, from metabolic functions to effects on the host immune system and transcriptome, can significantly impact patients’ dose requirements and even lead to toxicity[5,6].
Moreover, microbiome functioning may be altered by medicines in a patient-specific manner owing to the uniqueness of the microbiome composition[7,8]. Based on increasing evidence that drug–microbiome interactions can have clinically relevant effects on patient outcomes, it is important that healthcare professionals are aware of the findings and, where possible, account for them in their work.
Drug–microbiome relationships are relevant to all roles within the development and administration of medicines, including patient-facing clinicians, industry professionals, policy advisers and academic researchers. That said, assessment of such interactions does not typically feature in formal training or clinical guidelines. This may be due to the recentness of many findings and the large body of research remaining to be elucidated. Despite this, there are several ways in which healthcare professionals can immediately begin to consider the impact of the microbiome in their roles.
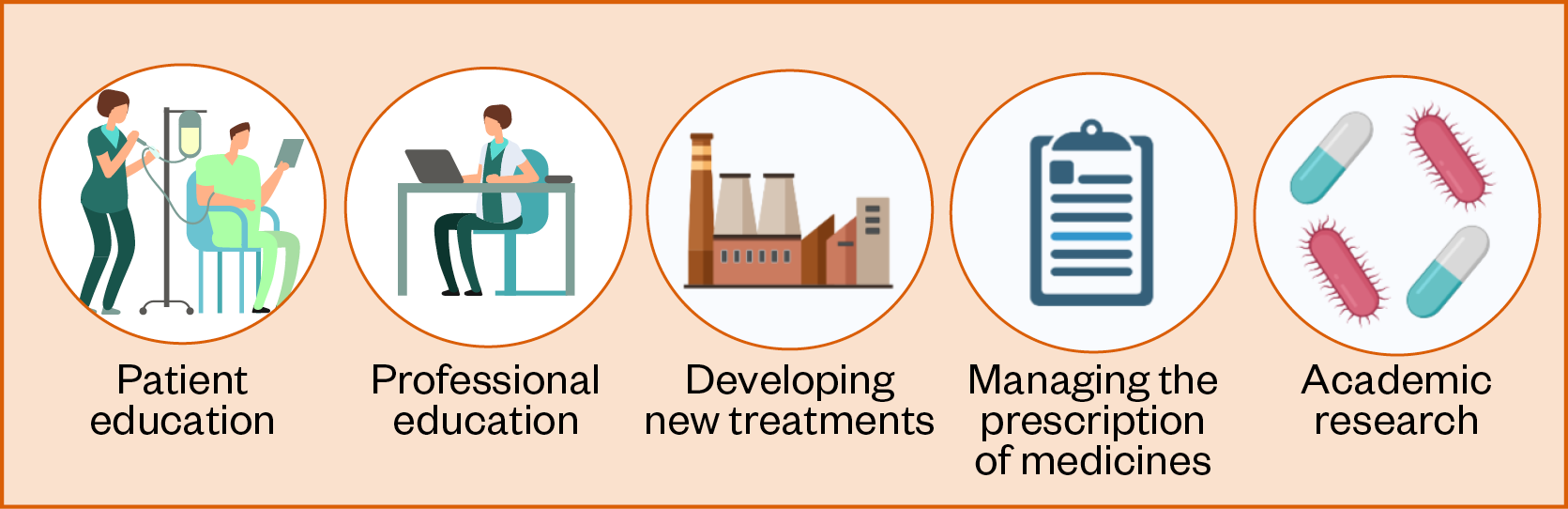
The second part of this review focuses on how healthcare professionals can ensure optimal management of drug–microbiome interactions when interacting with patients, managing the prescription of medicines, conducting research, educating others and developing new treatments (see Figure 1). However, an overview of the microbiome will first be provided, with respect to its relationships with human health and interactions with medicines. Important topics covered include: an up-to-date outline of documented pharmacomicrobiomic interactions; the effects of drugs on microbiome composition; the feasibility of basing clinical decisions on emerging evidence; and the benefits of screening for interactions in the preclinical phases of drug development.
The microbiome and its relationship with human health
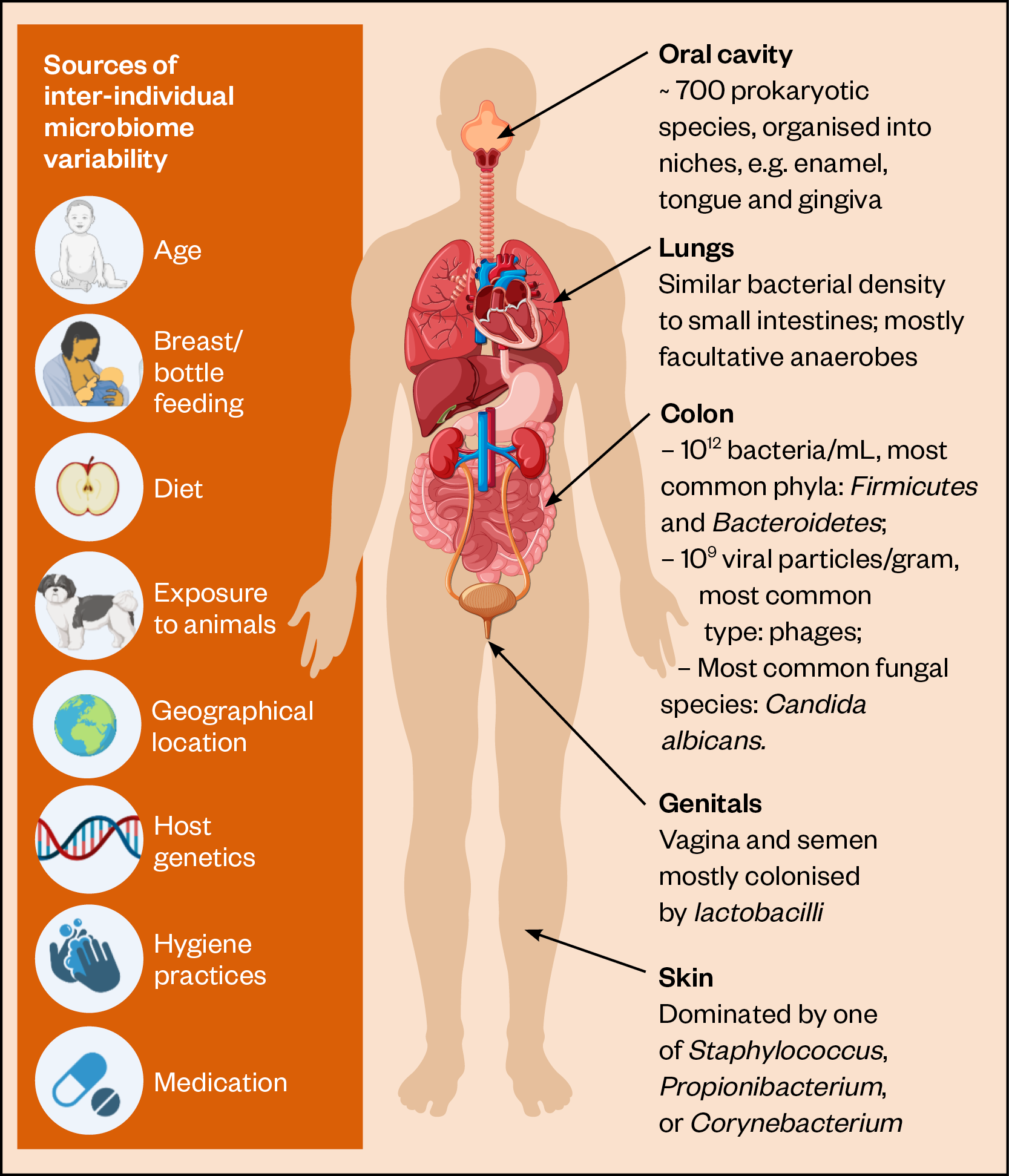
The human microbiome is composed of trillions of microorganisms, including the living microbiota (e.g. bacteria, fungi, and archaea) and non-living elements (e.g. eukaryotic viruses, phages, and plasmids)[9–11]. Microorganisms can be found on nearly all surfaces of the human body, including the skin, lungs, oral cavity and reproductive organs (see Figure 2).
The densest and most varied populations of microbes are found in the intestines, particularly in the colon[12]. In the colon, there are around 1 billion bacteria per mL content, each a member of up to a thousand possible species[13–15]. Within these species, there is also strain-level variation whereby microorganisms belonging to the same species differ genetically, often leading to functional differences; these differences can now be identified and investigated owing to advances in ‘omics’ technologies, such as shotgun metagenomic sequencing[16].
In the intestines, the gut microbiota is essential for the digestion of macronutrients, such as fibre; the production of vitamins and hormones; and the regulation of both immune function and gut epithelium integrity[17–19]. Elsewhere in the body, symbiotic microorganisms are involved in defending against sexually-transmitted infections, skin regeneration and protection against dental caries[20–22].
Over a lifetime, the microbiome composition changes in response to ageing and various external factors, such as diet, lifestyle, geography, cohabitation and medicine use[9,23,24]. Such changes can occur slowly, over years, or much more rapidly, over hours to days, owing to species variation[25]. To a lesser extent, host genetics can also play a role in microbiome composition, as shown by the gut microbiome in twin studies[26]. Owing to the complexity of the microbiome, and the many factors that influence it, the microorganisms that inhabit individuals are recognised to be as unique as a fingerprint[27].
Figure 2 shows significant elements of the microbiome at various body sites and sources of inter-individual microbiome variability[9,11,28–31].
Gut bacteria alone may encode up to 150 times more genes than their human hosts and the microbiome is now increasingly recognised as playing an intrinsic role in health[14,32,33]. Imbalance in the gastrointestinal microbiome, whereby species with pathogenic traits multiply to unhealthy concentrations, is a well known source of disease. Helicobacter pylori colonisation of the stomach and duodenum increases individuals’ risk of ulcers and cancer owing to chronic mucosal inflammation[34]. In addition, Clostridioides difficile (formerly Clostridium difficile), a bacterium that inhabits the colons of up to 90% of patients, can multiply to dangerous levels and produce toxins, often causing debilitating diarrhoea that can lead to colitis, sepsis and death[35].
The term ‘dysbiosis’ describes a state in which microbiome composition is imbalanced. Owing to imbalance in the number and types of microorganisms present at any particular body site, the microbiome can cease to perform its symbiotic functions, causing disease. Well-documented examples of diseases that may be triggered or worsened by dysbiosis include metabolic syndrome, autoimmune dysfunction, Parkinson’s disease and autism spectrum disorder[36–38]. Medication use can also be a significant trigger for dysbiosis[39,40]. While alteration of the microbiome composition may form part of a drug’s therapeutic action, in other cases, medicines can negatively impact the growth or functions of the gut microbiota. C. difficile infection is a prominent example of drug-induced dysbiosis because the disease is often instigated by the administration of broad-spectrum antibiotics[41]. Here, antibiotics substantially reduce intestinal bacterial diversity allowing the overgrowth of C. difficile. While antibiotic-induced C. difficile infection is a widely recognised condition, there are many other examples of microbiome-modulating drugs that are far less acknowledged[39].
The effect of drugs on the microbiome
Antimicrobials
Understandably, drugs aimed at reducing microbial growth can exert ruinous effects on the microbiome. Although intended to eradicate pathogenic microorganisms, antimicrobials administered via all routes are typically indiscriminate: killing or preventing the growth of both commensal and pathogenic species[42]. Antibiotics, antifungals and antivirals have the propensity to disrupt microbiome composition[39]. Most research to date has focused on the effects of antibiotics on the gut microbiome[43,44]. The extent to which an antimicrobial will affect the microbiome will depend on the drug, dosing and patient characteristics[45]. Antibiotic-induced gut microbiome changes can vary widely between and within individuals, depending on their age, sex, microbiome stability, and previous exposure to antimicrobials[25,46].
A study examining 12,422 full-term neonates found an association between antibiotic exposure during the first days after birth and higher body mass index (both sexes) and impaired weight and height gain (boys only), during the first six years of life[47]. These effects were attributed to significant alterations in the stool microbiome composition of a patient subset followed for 24 months after birth. Although these observations cannot amount to proof of causation, transfer of neonates’ faeces to germ-free mice did produce similar results (i.e. male mice administered faeces from antibiotic treated neonates had reduced weight gain compared to non-antibiotic treated controls)[47]. In adults, antibiotics can exert many effects on the gut microbiome, leading to depletion of vitamin-producing bacteria, reduced bacterial diversity and possibly increased risk of infection, inappropriate immune activity or altered digestion[44].
Antibiotic-induced dysbiosis can be longstanding, persisting years after treatment. Gut bacterial diversity may be lowered for up to four years following prescription of macrolides and lincosamides, and up to a year following prescriptions for beta-lactams and quinolones[40]. In addition, antimicrobial-induced changes may alter patients’ response to other pharmaceuticals; for example, antibiotics with considerable activity against intestinal Bacteroides fragilis have been found to increase bleeding risk in patients co-administered warfarin[48]. The bioavailability of the antipsychotic olanzapine may also be increased by antibiotic administration[49]. The use of broad-spectrum antibiotics may also alter immune response to vaccines; patients with low pre-existing antibodies against the H1N1 virus have exhibited impaired vaccine responses following a course of multiple oral antibiotics[50].
Human-targeted drugs
It may be surprising that drugs with human targets (i.e. with no intended antimicrobial action) can have multifarious effects on the microbiome. Many human targeted medicines, including oral and parenteral formulations, may alter microbiome composition and could impart wide-ranging consequences for patients. The possible scale of these effects can be appreciated from the results obtained in a 2018 study by Maier et al. where more than 1,000 drugs were incubated with 40 strains of gut bacteria and the drug’s effect on microbial growth measured.
Results from the in vitro experiment showed that 24% of human-targeted drugs significantly impaired the growth of at least one of the strains, with calcium channel blockers, antipsychotics and antineoplastics producing the highest antibacterial activity across all tested species. The findings from this work could indicate widespread and unexpected effects of medicines on patients’ gut microbiome composition; however, they should be taken in context of the experimental limitations.
First, bacteria were grown as monocultures (i.e. not in the presence of other microorganisms usually found in the intestines). It is known that bacteria function differently when growing within microbial communities compared to when alone; for example, owing to cross-feeding and competition from other microbes[51]. Therefore, Maier et al’s results may not be a totally accurate description of species’ response to drug exposure in an intestinal community.
Second, drug-bacteria incubations were measured for 16–24 hours[51]. In the intestinal environment, it is likely that most drugs would be absorbed into the systemic circulation long before the study endpoint; therefore, they may not have time to exert the same antibacterial effects observed in the study.
Several other researchers have now investigated the effects of drugs on the gut microbiome in vivo, providing data that are perhaps more clinically relevant than the high throughput in vitro screen conducted by Maier et al.[52].
Administration of atypical antipsychotics has been shown to significantly alter abundance of bacterial species associated with health (e.g. Akkermansia muciniphila) in adults of both sexes and decrease bacterial diversity in females, possibly indicating why females are more at risk of antipsychotic-induced weight gain[53]. Interestingly, while drug–microbiome effects could lead to dysbiosis, they could also form part of a drug’s mechanism of action. For instance, oral metformin administration has been found to increase the abundance of A. muciniphila and other beneficial species, possibly accounting for a portion of the drug’s positive effects on host metabolism[54]. In addition, statins seem to be protective against a gut microbiome composition that has been associated with systemic inflammation and obesity[55]. Further examples of drugs revealed to alter the gut microbiome in human or animal studies are fluoxetine, proton pump inhibitors, methotrexate, paracetamol, opioids and inhaled anticholinergics[56–59].
While these findings are interesting and uncover real alterations in microbiome structure, it is important to recognise that most current work reveals correlations between drugs and the microbiome rather than delineated mechanisms and causation. As such, to fully appreciate drugs’ effects on the microbiome, it is necessary to explore the in vivo interactions taking place and their clinical impact on patients. Technologies that may be useful for such investigations are causal inference networks, transcriptomics, proteomics and metabolomics[60].
The effect of the microbiome on drugs: pharmacomicrobiomics
Direct drug depletion by the microbiota
Humans have evolved alongside their microbiomes, and have come to rely on resident microorganisms to perform activities they cannot, from the digestion and absorption of food components to the detoxification of ingested poisons[17,61]. As such, the microbiota possesses considerable metabolic power. Most researched are the metabolic activities of gut bacteria, whose reactive potential has been compared to that of the liver[62]. Bacteria residing within the intestines produce enzymes that can chemically transform drugs, potentially altering PK and PD. In addition, certain strains of gut bacteria have recently been shown to accumulate drugs, in a process whereby the drug molecule is stored within the bacterial cell but not chemically modified[63,64].
Similar to biotransformation, bioaccumulation may decrease drug bioavailability, resulting in changes to PK. Together, drug transformation and accumulation represent the two methods by which the microbiota can deplete drug concentration within the intestinal lumen (see Figure 3)[63–70]
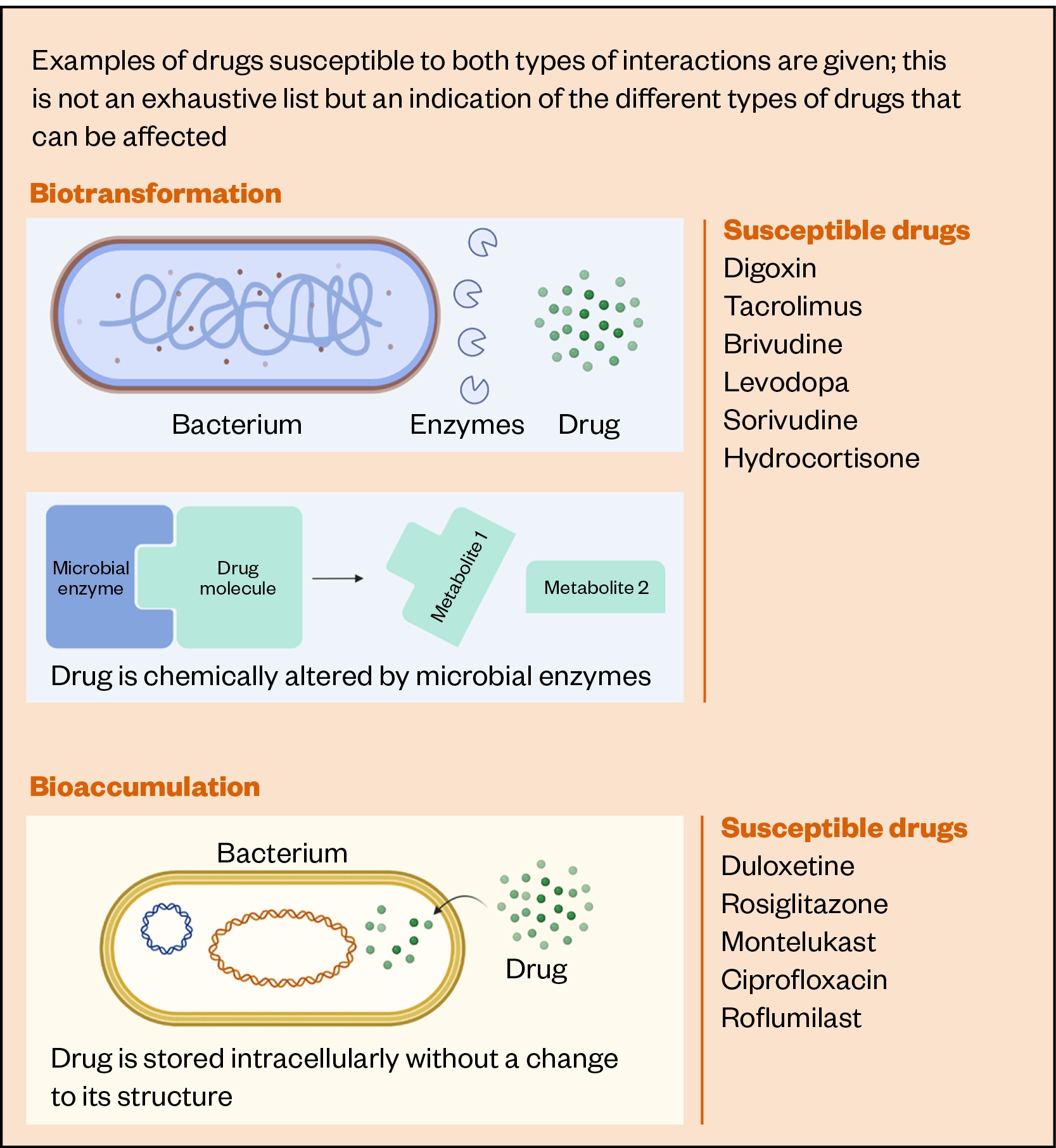
Biotransformation — the process by which organic compounds are transformed from one form to another, reducing the persistence and toxicity of the chemical compounds — has been reported as early as the 1930s, with the discovery that an early sulphonamide antibiotic (prontosil) was structurally activated by gut bacteria[71]. However, most biotransformation reactions have only been revealed in the past two decades[70,72–76]. Two high throughput in vitro screens, published in 2019 and 2020, have found more than 150 drugs to be significantly transformed by gut microbiota[70,76]. Drugs containing urea, azo, nitro and lactone groups were demonstrated to be particularly susceptible[76]. Further, analysis of interindividual variability found several drugs (ketoprofen, levonorgestrel, lovastatin, hydrocortisone and nicardipine) to be variably metabolised by the gut microbiota of 20 individuals[70].
Although not all these in vitro findings will translate to measurements in humans, a few drugs do now have validated microbial metabolism in human studies. For example, jejunal Enterococcus faecalis producing tyrosine decarboxylases can convert levodopa to dopamine, which can be subsequently converted to m-tyramine by dehydroxylase-producing Eggerthella lenta[66]. The presence of these bacterial enzymes predicts levodopa metabolism in people with Parkinson’s disease, leading to increased dosing requirements[77].
Similarly, patients who have undergone kidney transplant with high stool abundances of Faecalibacterium prausnitzii have been observed to require higher doses of tacrolimus, owing to the drug’s biotransformation into a metabolite with 15-fold lower immunosuppressant activity[5,67]. In some cases, biotransformation may not only alter a drug’s PK, but could alter PD, as microbial drug metabolites could exert unexpected physiological activities. For example, the antiviral sorivudine undergoes microbial hydrolysis to bromovinyluracil and subsequent host metabolism to an inhibitor of dihydropyrimidine dehydrogenase, a hepatic enzyme[68]. Tragically, this reaction was only identified after the death of 18 oncology patients in 1993, who were co-administered sorivudine and 5-fluorouracil (5-FU)[68]. Here, the inhibition of dihydropyrimidine dehydrogenase led to the accumulation of 5-FU in patients, resulting in diarrhoea, pancytopenia and eventual death[68]. This highlights the importance of screening for drug–microbiome interactions before drugs are administered to patients.
In comparison to biotransformation, there is much less evidence regarding the microbial accumulation of drugs. A 2021 study by Klünemann et al. published 17 cases of bioaccumulation, which were measured by incubating 12 small molecule drugs with 25 strains of gut bacteria (resulting in 375 bacteria-drug combinations)[63]. Here, duloxetine, montelukast, rosiglitazone, and roflumilast were identified as accumulation-susceptible drugs. Elsewhere, the accumulation of 100 antibiotics by a strain of Escherichia coli was measured, revealing that antibiotics with low globularity, high amphiphilicity and rigidity, and containing an amine group, were most likely to be accumulated[64]. This work also identified that porin channels represented key routes for drugs’ traversal of the bacterial outer membrane.
Indirect pharmacokinetic effects
Direct metabolism by the microbiota is not the only way that the microbiome can alter drugs’ PK. The microbiome can affect drugs’ absorption, metabolism and excretion via indirect mechanisms. First, intestinal drug absorption may be altered by changes to epithelial permeability, gut motility, intestinal drug transporters and bile acids (see Figure 4).
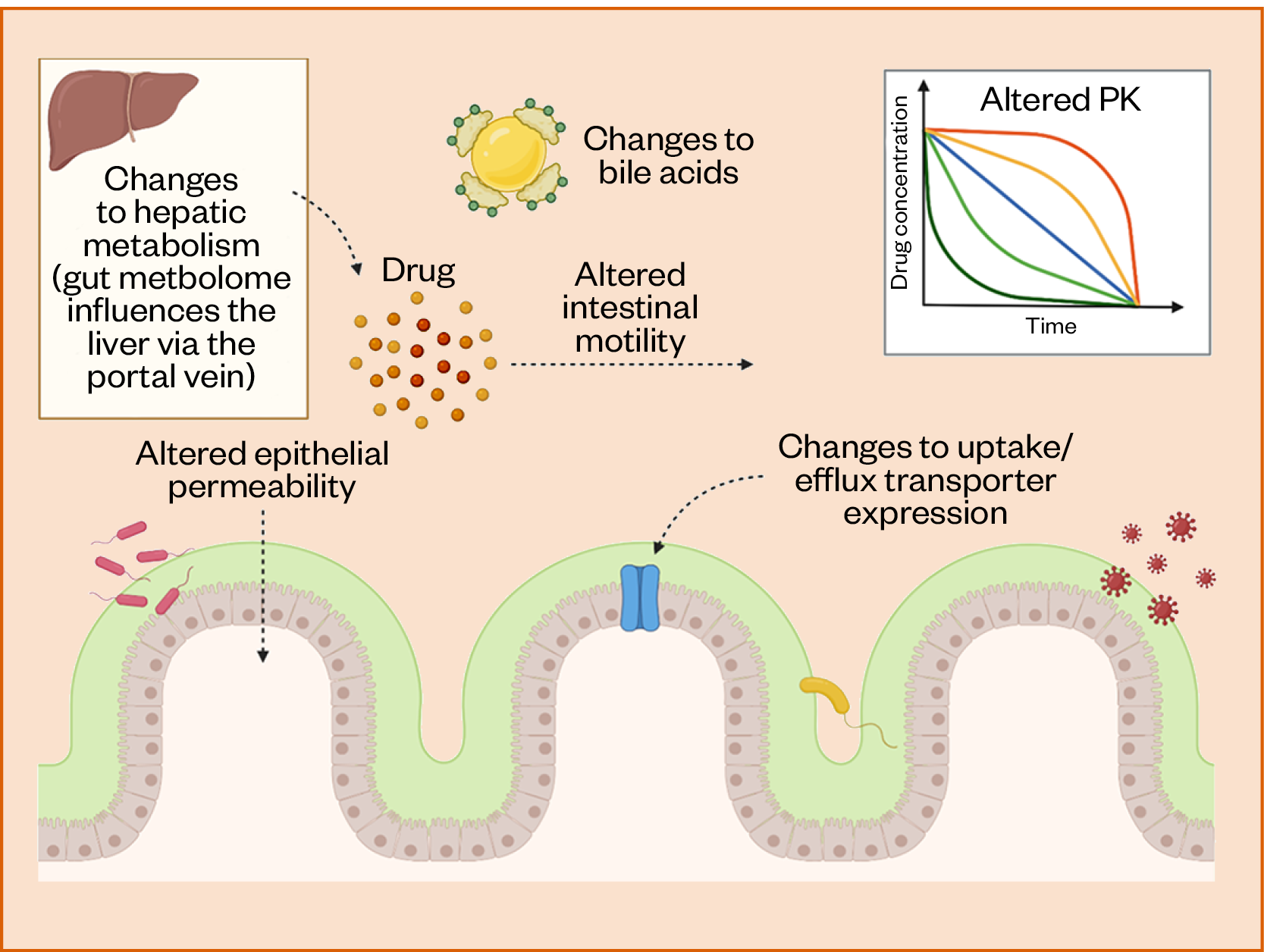
The gut microbiome is an important modulator of epithelial tight junction integrity, acting via immunoregulatory pathways[78–81]. This is also true at other body sites, such as the skin[82]. In dysbiotic states, an unbalanced microbiota can cease to maintain barrier integrity, potentially leading to increased drug absorption[83–85]. The expression of epithelial drug efflux transporters can also be affected by microbiome composition, altering the transport of drugs over the gut epithelium[86]. In addition, drug absorption may be affected by microbiome effects on intestinal motility. The richness and diversity of colonic bacteria is positively correlated with colonic transit time[87]. Therefore, patients with more diverse colonic microbiomes may absorb drugs in delayed-release formulations to a greater extent than patients with less microbial diversity, owing to longer colonic transits.
Modifications to bile acids, which play a role in lipophilic drug solubilisation, can also be triggered by the metabolic activities of the microbiota in the distal gut[88,89]. Such changes have been found to impact the solubility capacity of nine oral drugs, including phenytoin, which raises concerns owing to the drug’s critical indications and narrow therapeutic index[90].
Hepatic drug metabolites excreted in bile can be ‘reactivated’ by colonic bacteria, extending their systemic half-lives, in a process known as enterohepatic reabsorption[91]. Furthermore, drug metabolism in the liver may be significantly altered by changes to gut microbiome composition, owing to the transfer of microbial metabolites from the intestine to the liver via the hepatic portal vein, and subsequent effects on the hepatic transcriptome. Results from a 2016 study revealed more than 4,000 genes to be differentially expressed in the livers of germ-free and colonised mice, several of which were implicated in drug metabolism, such as Cyp2b10 and Cyp3a11[92]. It is important to recognise that these results may not be fully transferable to humans; however, the work does indicate a potentially important relationship between the gut microbiota and hepatic metabolism. Certain diseases of the liver, such as alcoholic hepatitis, have been associated with substantial changes to the gut metagenome and metabolome, suggesting that microbiome-liver relationships are likely present in human patients[93].
Patient education
The public are developing an increasing interest in microbiome health. Scientific development describing new disease-microbiome associations and microbiome-targeted medicines are reaching mainstream news, direct-to-consumer microbiome sequencing kits have been developed and sales of probiotics and fermented foods are experiencing a surge in popularity[94–97]. As such, it is increasingly likely that patient-facing healthcare professionals will be asked questions relating to interactions between medicines and the microbiome; for example, the risk of a medicine causing dysbiosis, the best probiotic product to take for a given indication, or how drug-microbiome interactions could affect a treatment’s therapeutic efficacy or toxicity (see Box[50,52,98,99]).
Box 1: Potential questions that patients may ask healthcare professionals relating to drug-microbiome interactions
- Am I at risk of dysbiosis?
- Should I take probiotics with antibiotics?
- Will this medicine damage my microbiome?
- Will my microbiome affect my treatment success?
- Could my microbiome alter my vaccine response?
When speaking to patients about drugs’ effects on the microbiome, it is important to convey the current infancy of the field. While several drugs have been implicated in altering microbiome composition, it is often not clear how likely alterations are to impact specific patients’ health. In cases where evidence points to medicines having positive effects on the microbiome, professionals may wish to communicate this evidence as part of wider patient counselling to potentially promote medication adherence. If professionals suspect medicines may impact a patient’s microbiome negatively, they may wish to discuss this risk with the wider healthcare team and the patient to manage the risk of dysbiosis alongside the clinical picture.
Some patients may also enquire as to the efficacy and/or suitability of probiotic formulations, such as preparations containing Lactobacillus strains, for a given indication. As over-the-counter probiotic products are currently not subject to the same stringent regulations as drug products, this can be a difficult query to answer, as the efficacy of many probiotics has not been validated. Several factors can influence the ability of a probiotic product to effectively exert a therapeutic benefit, including the identity and concentration of microbial strains, the formulation type, the level of quality control during manufacturing, and the product stability[100].
Formulations should enable the safe passage of probiotic strains through the stomach, as exposure to gastric acid can substantially reduce microbial viability. A 2015 study measuring the in vitro acid tolerance of eight commercial probiotic formulations found that liquid-based products promoted probiotic viability to a greater extent than freeze-dried products[101]. Solid oral dosage forms may be protected through the use of coatings designed to resist degradation in the upper gastrointestinal tract[102].
A common reason for patients to seek a probiotic product is the recent administration of antibiotics, to limit the chance of ensuing dysbiosis. Although this practice is frequent, it is not necessarily evidence based. A study examining human gut microbiome recovery after broad spectrum antibiotic exposure found that an enteric coated probiotic (containing at least 25 billion bacteria from several strains, mainly lactobacilli and bifidobacteria) actually impaired microbiome reconstitution[99].
On the other hand, a large meta-analysis found that administration of probiotics (strains varied between studies) significantly reduced patients’ risk of antibiotic-associated diarrhoea: a common symptom of intestinal dysbiosis[103]. Since antimicrobial-associated dysbiosis is highly variable, it is likely that there is no ‘perfect’ probiotic that can prevent or treat dysbiosis in all patients. It is likely that precision probiotics, composed of microbial strains personalised to individual cases, will produce more consistent benefits[104]. Rather than prescribing a generic mix of probiotics to prevent dysbiosis, it is probable that carefully selected species with characterised functionalities will be more efficacious[105]. While precision probiotics are not currently a reality, advances in microbiome science mean that they may soon reach clinical practice[98]. Identification of the most suitable probiotic strains for specific patients will likely require metagenomic sequencing to identify the compositional and/or functional deficits in individual microbiomes.
Communicating the impact of pharmacomicrobiomics to patients can be challenging. For one, there is a stark lack of evidence concerning interactions involving microorganisms outside of the intestines. For example, the effect of the skin microbiome on the PK and PD of topical drugs is underexplored. Furthermore, with a few exceptions, the impact of drug–gut microbiome interactions on patients’ dose requirements is largely unknown. Therefore, the clinical relevance of reactions measured using in vitro and animal models can be difficult to interpret.
It is expected that the clinical impact of pharmacomicrobiomics will be increasingly characterised as more human studies are conducted. With this research, healthcare professionals may be given the tools to predict drug–microbiome interactions on an individual basis. This could involve the use of point-of-care tests that measure microbiome composition via sequencing and then infer how an individual’s results could influence their dose requirements. For example, determination of a patient’s stool F. prausnitzii abundance could guide their starting dose of tacrolimus[5]. If this becomes a reality, then healthcare professionals should be comfortable in explaining the drug–microbiome interactions underpinning the tests to patients. In addition, healthcare professionals using such tests should be confident that underlying evidence is applicable to their patient. For instance, dose–microbiome correlations gathered from certain patient groups may not always be relevant to other patient groups.
Managing the prescription of medicines
As evidence continues to emerge, it is likely that professionals will be required to increasingly incorporate drug-microbiome interactions into prescribing guidelines. At present, guidelines already exist around preventing antibiotic-induced C. difficile infection, which include measures such as: avoiding repeat prescriptions of antimicrobials; considering non-antimicrobial interventions; identifying sources of infection; considering local resistance patterns; and prescribing the shortest effective course via the most appropriate dose and route when necessary[106].
As C. difficile infection is an outcome of gut dysbiosis, these existing guidelines provide good advice for protection against dysbiosis in general. It perhaps owes to the overt and immediately life-threatening nature of C. difficile infection that this consequence of dysbiosis has received substantial attention whereas others have not[41]. As associated evidence is hopefully strengthened over the coming years, the effects of antibiotic-induced dysbiosis on other pharmaceuticals, such as warfarin, olanzapine, and vaccines, may be integrated into prescribing guidance[48–50]. Furthermore, the impact of non-antimicrobial drugs on the microbiome may be more considered. For instance, the prescription of proton pump inhibitors is now cautioned in patients at risk of gastrointestinal infections (including C. difficile), following reports that their pH-raising effects promotes microbial growth in the stomach[107].
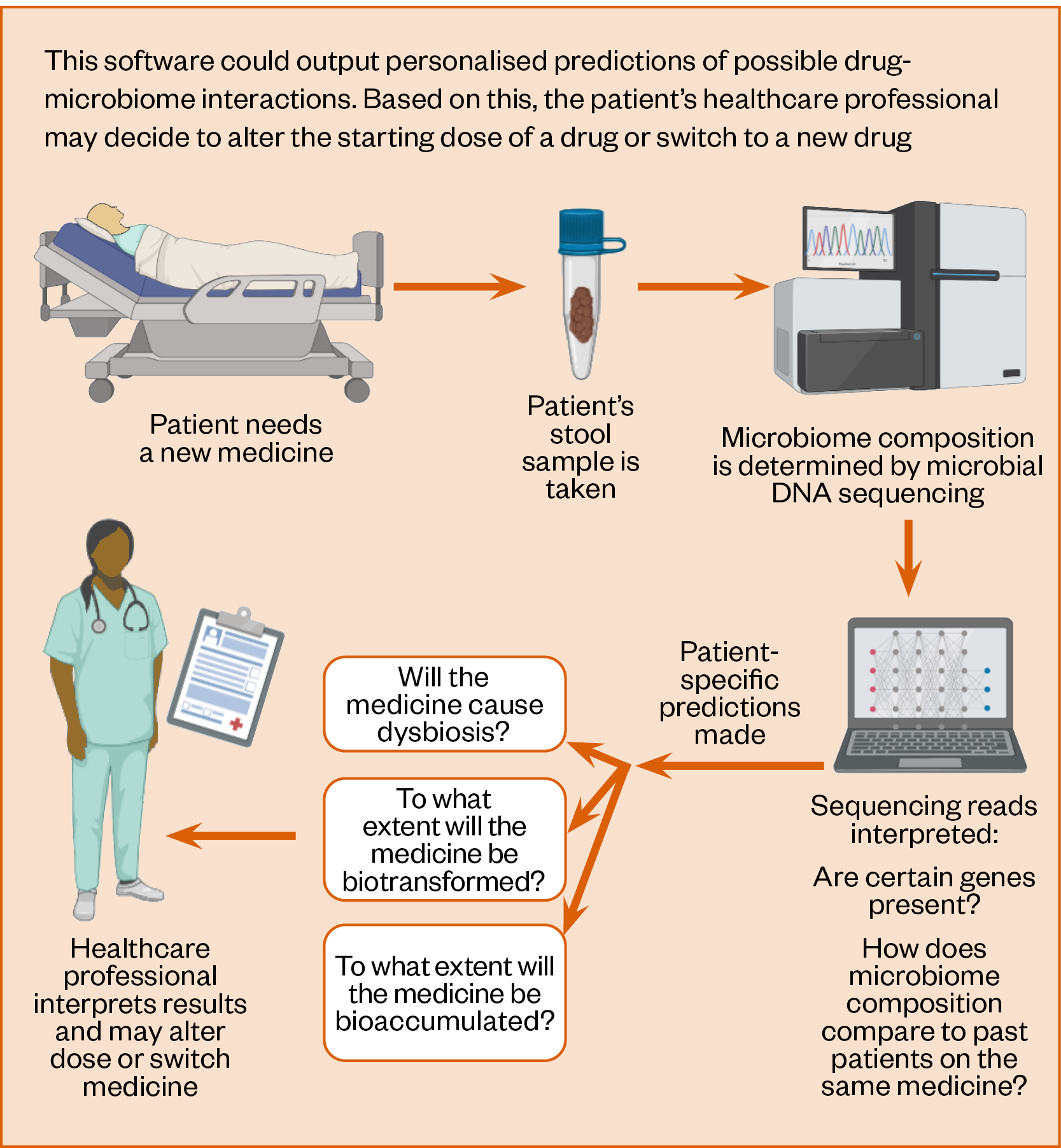
Microbiome sequencing is becoming progressively cheaper, faster, and more accurate. Consequently, the characterisation of patients’ microbiomes may soon influence the prescribing process (see Figure 5). Just as it is second nature for clinicians to check a patient’s creatinine clearance, liver function tests or blood pressure before initiating a new medicine, this may become the norm for microbiome composition. Information on patients’ microbiome may allow professionals to predict individual risk of drug-microbiome interactions. This could facilitate the identification of patients at risk of drug-induced dysbiosis or microbial drug depletion.
Such knowledge could prompt healthcare professionals to adjust the doses of drugs or even switch to alternative therapeutics. To enable this, software that can relate raw sequencing reads to probability of drug-microbiome interactions will be necessary. To date, substantial efforts have been made to realise this outcome, with enabling technologies, such as machine learning (ML), receiving significant attention.
In 2018, Mallory et al. used unsupervised ML to predict drugs’ metabolism by bacterial enzymes using reaction vectors[108]. Here, vectors were composed of known small molecule–bacterial enzyme reactions and untested drugs’ probability of reaction was computed using ML clustering.
Zimmerman et al. also utilised clustering in their drug–bacteria screening study to identify phyla-specific metabolic activities and the functional groups that increased drugs’ susceptibility to depletion[76]. Elsewhere, supervised ML has been employed to predict drugs’ effects on the gut microbiota’s growth and whether drugs are at risk of biotransformation or bioaccumulation[109,110]. These studies represent progress within the field, as they have successfully related drug structure to probability of interactions with the gut microbiome.
To advance further, research could consider both drug structures and the genetic sequence of microorganisms to characterise how strain-level characteristics affect interactions with drugs’ chemical features. Moreover, more work is needed to understand the mechanisms underlying the bioaccumulation of drugs, and the occurrence of drug-microbiome interactions outside of the gut. Finally, these capabilities should be combined to predict the likelihood and clinical impact of drug-microbiome interactions in real patients, a task that will require analysis of large patient cohorts. In the field of nutrition, the metagenomic sequences of 1,908 individuals’ faecal microbiomes were used to predict postprandial metabolic response, demonstrating the promise of similar methodologies for the field of pharmacomicrobiomics[23].
Academic research and education
There is great opportunity for university researchers to contribute towards emerging drug–microbiome evidence. At present, there remains a significant amount of work to be done in characterising the drug–microbiome axis. Important areas in need of greater research include the mechanistic description of interactions, measurement of interactions’ clinical impact, prediction of interactions’ occurrence in individuals and analysis of interactions outside of the gut. Projects exploring these areas will likely require a multidisciplinary approach, involving researchers with skills such as microbiology, pharmacometabolomics, genomics, in silico modelling and the organisation of studies involving patients.
While researchers working within academic settings are expected to conduct most of the fundamental science, there will be increasing opportunity for those working within clinical settings to collaborate on projects involving patients. Professionals within healthcare settings could also contribute to evidence through audits and data analysis projects, whereby possible interactions between drugs and the microbiome could be identified from medical records. Medical records have already been utilised to identify an association between use of antibiotics with high B. fragilis activity and extended prothrombin time in patients taking warfarin[48]. Such work could provide important indicators of clinically relevant drug–microbiome interactions that could subsequently be mechanistically explored in laboratories.
To fully spread awareness of drug–microbiome relationships, it is important to educate current and future healthcare professionals. Teaching can be incorporated into existing training modules, such as those focusing on PK, PD and toxicology. Inspiration may be taken from other specialities, such as public health, where microbiome science is being taught through both dedicated courses and as part of more general modules[111].
Developing new treatments
Upon discovery, a novel drug candidate will undergo stringent preclinical and clinical testing prior to market approval. Owing to the risk and investment involved, only 1 in 10,000 candidate molecules are typically advanced from preclinical to clinical trials; compounds must perform well across all testing categories[112]. Despite the risk that drug–microbiome interactions pose to a compound’s PK variability and toxicity profile, they are not routinely tested for during drug development[113].
As demonstrated by the sorivudine tragedy, early screening for drug–microbiome interactions is highly advisable[68]. Although documented cases of microbiome-mediated drug toxicity are rare, microbial metabolism of drugs has been implicated in promoting PK variability between patients[5,77]. This PK variability could cause problems for companies during clinical trials, as treatments may not achieve their clinical endpoints if patients do not respond to the treatment uniformly. The primary benefit of identifying a new drug’s interactions with the microbiome is that risks to patients can be assessed well before the clinical phase begins. In addition, companies could save significant funds by ending the progression of unsuitable drugs.
Screening for drug-microbiome interactions could involve the use of the human faecal microbiota, animal models, in silico methods, or high throughput in vitro analyses[74,75,114]. Important questions to ask include: ‘Is this drug’s PK or PD affected by the microbiome?’ and ‘Could this drug alter microbiome composition?’. Results showing PK and PD effects may highlight potential dosing challenges in clinical phases, owing to the wide variability between patients’ microbiomes.
In addition, if investigational drugs are found to alter microbiome composition, then investigations into the risk of dysbiosis — or even repurposing opportunities — could be conducted. For example, a drug may have unexpected beneficial effects on microbiome composition that could be transferred to other indications[115]. To save laboratory resources, in silico prediction of new drugs’ interactions with the microbiome could be incorporated into the drug discovery phase[52]. Here, existing ML algorithms with reliable performances could begin to be used immediately[108–110]. The chemical features that promote microbial metabolism could also be considered in the design of new drugs, through awareness of functional groups that increase risk of biotransformation or bioaccumulation[64,76].
Prediction of a drug’s stability in the presence of the microbiota could also inform formulation strategies. For example, work conducted by AstraZeneca, published in 2014, found a strong correlation (R2=0.9) between drugs’ permeability through the human colonic epithelium and their stability in the presence of the faecal microbiota[116]. If a new active compound is identified as being depleted by the microbiota at a particular site within the intestines, then targeted delivery of the compound to an alternative site could protect its bioavailability. Site specific delivery of oral drugs can be achieved using coatings designed to selectively release drugs in defined regions of the gastrointestinal tract[117]. As examples, two polysaccharide-based coating technologies have recently been launched on to the market for colonic drug delivery[118,119]. Both systems combine resistant starch and a pH sensitive polymer to achieve fail-safe delivery of any drug to the colon, by exploiting intestinal changes in microbial concentrations and pH[120].
Conclusions
Research over the past 20 years has revealed the importance of the human microbiome for health and uncovered many ways it can interact with medicines. Drug–microbiome relationships can be bidirectional: drugs can affect the microbiome, and the microbiome can affect drugs’ PK and PD, both directly and indirectly. Since microbiome composition is unique to individuals and changes in response to numerous factors, it is currently difficult to predict drug–microbiome interactions for specific patients. Despite this, it is likely that healthcare professionals will be increasingly called upon to incorporate the drug–microbiome relationship in their work.
Patients may ask whether they should take probiotics with their antibiotics, prescribers may query which drugs are least likely to cause dysbiosis in their patients, and drafting of new prescribing guidelines may require consideration of drug-microbiome interactions. While personalised predictions of drug–microbiome interactions are not yet a clinical reality, those writing and screening prescriptions are well placed to advise on the current best evidence. Professionals working in research and drug development can contribute towards emerging evidence by identifying and characterising new drug–microbiome interactions; information that can eventually inform prescribing guidelines in the clinic.
In addition, as healthcare professionals are trained, it is essential that they are educated on the importance of drug–microbiome relationships. In coming years, it is a distinct possibility that microbiome profiling will become a common clinical investigation in healthcare settings. If so, software enabling clinicians to generate personalised predictions will likely be developed, allowing them to consider drug–microbiome interactions at the individual patient level.
Financial disclosure/conflict of interest statement
Abdul W Basit has been granted patents relating to the OPTICORE™ and Phloral® technologies.
This work was supported by The Engineering and Physical Sciences Research Council (EPSRC) under Grant [EP/S023054/1].
- 1Vinarov Z, Abdallah M, Agundez JAG, et al. Impact of gastrointestinal tract variability on oral drug absorption and pharmacokinetics: An UNGAP review. European Journal of Pharmaceutical Sciences. 2021;162:105812. doi:10.1016/j.ejps.2021.105812
- 2Sharma A, Buschmann MM, Gilbert JA. Pharmacomicrobiomics: The Holy Grail to Variability in Drug Response? Clin. Pharmacol. Ther. 2019;106:317–28. doi:10.1002/cpt.1437
- 3Doestzada M, Vila AV, Zhernakova A, et al. Pharmacomicrobiomics: a novel route towards personalized medicine? Protein Cell. 2018;9:432–45. doi:10.1007/s13238-018-0547-2
- 4Beger RD, Schmidt MA, Kaddurah-Daouk R. Current Concepts in Pharmacometabolomics, Biomarker Discovery, and Precision Medicine. Metabolites. 2020;10:129. doi:10.3390/metabo10040129
- 5Lee JR, Muthukumar T, Dadhania D, et al. Gut Microbiota and Tacrolimus Dosing in Kidney Transplantation. PLoS ONE. 2015;10:e0122399. doi:10.1371/journal.pone.0122399
- 6Chaput N, Lepage P, Coutzac C, et al. Baseline gut microbiota predicts clinical response and colitis in metastatic melanoma patients treated with ipilimumab. Annals of Oncology. 2017;28:1368–79. doi:10.1093/annonc/mdx108
- 7Ramakrishna C, Mendonca S, Ruegger PM, et al. Herpes simplex virus infection, Acyclovir and IVIG treatment all independently cause gut dysbiosis. PLoS ONE. 2020;15:e0237189. doi:10.1371/journal.pone.0237189
- 8Hou Q, Zhao F, Liu W, et al. Probiotic-directed modulation of gut microbiota is basal microbiome dependent. Gut Microbes. 2020;12:1736974. doi:10.1080/19490976.2020.1736974
- 9Liang G, Bushman FD. The human virome: assembly, composition and host interactions. Nat Rev Microbiol. 2021;19:514–27. doi:10.1038/s41579-021-00536-5
- 10Borrel G, Brugère J-F, Gribaldo S, et al. The host-associated archaeome. Nat Rev Microbiol. 2020;18:622–36. doi:10.1038/s41579-020-0407-y
- 11Structure, function and diversity of the healthy human microbiome. Nature. 2012;486:207–14. doi:10.1038/nature11234
- 12Lloyd-Price J, Abu-Ali G, Huttenhower C. The healthy human microbiome. Genome Med. 2016;8. doi:10.1186/s13073-016-0307-y
- 13Martinez-Guryn K, Leone V, Chang EB. Regional Diversity of the Gastrointestinal Microbiome. Cell Host & Microbe. 2019;26:314–24. doi:10.1016/j.chom.2019.08.011
- 14Qin J, Li R, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi:10.1038/nature08821
- 15Zou Y, Xue W, Luo G, et al. 1,520 reference genomes from cultivated human gut bacteria enable functional microbiome analyses. Nat Biotechnol. 2019;37:179–85. doi:10.1038/s41587-018-0008-8
- 16Van Rossum T, Ferretti P, Maistrenko OM, et al. Diversity within species: interpreting strains in microbiomes. Nat Rev Microbiol. 2020;18:491–506. doi:10.1038/s41579-020-0368-1
- 17Tian L, Wang X-W, Wu A-K, et al. Deciphering functional redundancy in the human microbiome. Nat Commun. 2020;11. doi:10.1038/s41467-020-19940-1
- 18James KR, Gomes T, Elmentaite R, et al. Distinct microbial and immune niches of the human colon. Nat Immunol. 2020;21:343–53. doi:10.1038/s41590-020-0602-z
- 19Heintz-Buschart A, Wilmes P. Human Gut Microbiome: Function Matters. Trends in Microbiology. 2018;26:563–74. doi:10.1016/j.tim.2017.11.002
- 20Bullington BW, Lee MH, Mlingi J, et al. Cervicovaginal bacterial communities in reproductive-aged Tanzanian women with Schistosoma mansoni, Schistosoma haematobium, or without schistosome infection. ISME J. 2021;15:1539–50. doi:10.1038/s41396-020-00868-9
- 21Wang G, Sweren E, Liu H, et al. Bacteria induce skin regeneration via IL-1β signaling. Cell Host & Microbe. 2021;29:777-791.e6. doi:10.1016/j.chom.2021.03.003
- 22Freire M, Moustafa A, Harkins DM, et al. Longitudinal Study of Oral Microbiome Variation in Twins. Sci Rep. 2020;10. doi:10.1038/s41598-020-64747-1
- 23Asnicar F, Berry SE, Valdes AM, et al. Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals. Nat Med. 2021;27:321–32. doi:10.1038/s41591-020-01183-8
- 24Wilmanski T, Diener C, Rappaport N, et al. Gut microbiome pattern reflects healthy ageing and predicts survival in humans. Nat Metab. 2021;3:274–86. doi:10.1038/s42255-021-00348-0
- 25Lozupone CA, Stombaugh JI, Gordon JI, et al. Diversity, stability and resilience of the human gut microbiota. Nature. 2012;489:220–30. doi:10.1038/nature11550
- 26Goodrich JK, Davenport ER, Beaumont M, et al. Genetic Determinants of the Gut Microbiome in UK Twins. Cell Host & Microbe. 2016;19:731–43. doi:10.1016/j.chom.2016.04.017
- 27Franzosa EA, Huang K, Meadow JF, et al. Identifying personal microbiomes using metagenomic codes. Proc Natl Acad Sci USA. 2015;112:E2930–8. doi:10.1073/pnas.1423854112
- 28Strati F, Di Paola M, Stefanini I, et al. Age and Gender Affect the Composition of Fungal Population of the Human Gastrointestinal Tract. Front. Microbiol. 2016;7. doi:10.3389/fmicb.2016.01227
- 29Moffatt MF, Cookson WO. The lung microbiome in health and disease. Clin Med. 2017;17:525–9. doi:10.7861/clinmedicine.17-6-525
- 30Gao L, Xu T, Huang G, et al. Oral microbiomes: more and more importance in oral cavity and whole body. Protein Cell. 2018;9:488–500. doi:10.1007/s13238-018-0548-1
- 31Koedooder R, Mackens S, Budding A, et al. Identification and evaluation of the microbiome in the female and male reproductive tracts. Human Reproduction Update. 2018;25:298–325. doi:10.1093/humupd/dmy048
- 32The Integrative Human Microbiome Project. Nature. 2019;569:641–8. doi:10.1038/s41586-019-1238-8
- 33Lynch SV, Pedersen O. The Human Intestinal Microbiome in Health and Disease. N Engl J Med. 2016;375:2369–79. doi:10.1056/nejmra1600266
- 34Dunn BE, Cohen H, Blaser MJ. Helicobacter pylori. Clin Microbiol Rev. 1997;10:720–41. doi:10.1128/cmr.10.4.720
- 35Furuya-Kanamori L, Marquess J, Yakob L, et al. Asymptomatic Clostridium difficile colonization: epidemiology and clinical implications. BMC Infect Dis. 2015;15. doi:10.1186/s12879-015-1258-4
- 36Singer-Englar T, Barlow G, Mathur R. Obesity, diabetes, and the gut microbiome: an updated review. Expert Review of Gastroenterology & Hepatology. 2018;13:3–15. doi:10.1080/17474124.2019.1543023
- 37Markle JGM, Frank DN, Mortin-Toth S, et al. Sex Differences in the Gut Microbiome Drive Hormone-Dependent Regulation of Autoimmunity. Science. 2013;339:1084–8. doi:10.1126/science.1233521
- 38Cryan JF, O’Riordan KJ, Sandhu K, et al. The gut microbiome in neurological disorders. The Lancet Neurology. 2020;19:179–94. doi:10.1016/s1474-4422(19)30356-4
- 39Maier L, Pruteanu M, Kuhn M, et al. Extensive impact of non-antibiotic drugs on human gut bacteria. Nature. 2018;555:623–8. doi:10.1038/nature25979
- 40Mulder M, Radjabzadeh D, Kiefte-de Jong JC, et al. Long-term effects of antimicrobial drugs on the composition of the human gut microbiota. Gut Microbes. 2020;12:1791677. doi:10.1080/19490976.2020.1791677
- 41Fadda HM. The Route to Palatable Fecal Microbiota Transplantation. AAPS PharmSciTech. 2020;21. doi:10.1208/s12249-020-1637-z
- 42Baquero F, Levin BR. Proximate and ultimate causes of the bactericidal action of antibiotics. Nat Rev Microbiol. 2020;19:123–32. doi:10.1038/s41579-020-00443-1
- 43Pérez-Cobas AE, Gosalbes MJ, Friedrichs A, et al. Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut. 2012;62:1591–601. doi:10.1136/gutjnl-2012-303184
- 44Langdon A, Crook N, Dantas G. The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 2016;8. doi:10.1186/s13073-016-0294-z
- 45Jernberg C, Löfmark S, Edlund C, et al. Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology. 2010;156:3216–23. doi:10.1099/mic.0.040618-0
- 46Gao H, Shu Q, Chen J, et al. Antibiotic Exposure Has Sex-Dependent Effects on the Gut Microbiota and Metabolism of Short-Chain Fatty Acids and Amino Acids in Mice. mSystems. 2019;4. doi:10.1128/msystems.00048-19
- 47Uzan-Yulzari A, Turta O, Belogolovski A, et al. Neonatal antibiotic exposure impairs child growth during the first six years of life by perturbing intestinal microbial colonization. Nat Commun. 2021;12. doi:10.1038/s41467-020-20495-4
- 48Yagi T, Naito T, Kato A, et al. Association Between the Prothrombin Time–International Normalized Ratio and Concomitant Use of Antibiotics in Warfarin Users: Focus on Type of Antibiotic and Susceptibility of Bacteroides fragilis to Antibiotics. Ann Pharmacother. 2020;55:157–64. doi:10.1177/1060028020940728
- 49Cussotto S, Walsh J, Golubeva AV, et al. The gut microbiome influences the bioavailability of olanzapine in rats. EBioMedicine. 2021;66:103307. doi:10.1016/j.ebiom.2021.103307
- 50Hagan T, Cortese M, Rouphael N, et al. Antibiotics-Driven Gut Microbiome Perturbation Alters Immunity to Vaccines in Humans. Cell. 2019;178:1313-1328.e13. doi:10.1016/j.cell.2019.08.010
- 51Goyal A, Wang T, Dubinkina V, et al. Ecology-guided prediction of cross-feeding interactions in the human gut microbiome. Nat Commun. 2021;12. doi:10.1038/s41467-021-21586-6
- 52McCoubrey LE, Gaisford S, Orlu M, et al. Predicting drug-microbiome interactions with machine learning. Biotechnology Advances. 2022;54:107797. doi:10.1016/j.biotechadv.2021.107797
- 53Flowers SA, Evans SJ, Ward KM, et al. Interaction Between Atypical Antipsychotics and the Gut Microbiome in a Bipolar Disease Cohort. Pharmacotherapy. 2017;37:261–7. doi:10.1002/phar.1890
- 54Wu H, Esteve E, Tremaroli V, et al. Metformin alters the gut microbiome of individuals with treatment-naive type 2 diabetes, contributing to the therapeutic effects of the drug. Nat Med. 2017;23:850–8. doi:10.1038/nm.4345
- 55Vieira-Silva S, Falony G, et al. Statin therapy is associated with lower prevalence of gut microbiota dysbiosis. Nature. 2020;581:310–5. doi:10.1038/s41586-020-2269-x
- 56Fung TC, Vuong HE, Luna CDG, et al. Intestinal serotonin and fluoxetine exposure modulate bacterial colonization in the gut. Nat Microbiol. 2019;4:2064–73. doi:10.1038/s41564-019-0540-4
- 57Jackson MA, Verdi S, Maxan M-E, et al. Gut microbiota associations with common diseases and prescription medications in a population-based cohort. Nat Commun. 2018;9. doi:10.1038/s41467-018-05184-7
- 58Freedberg DE, Toussaint NC, Chen SP, et al. Proton Pump Inhibitors Alter Specific Taxa in the Human Gastrointestinal Microbiome: A Crossover Trial. Gastroenterology. 2015;149:883-885.e9. doi:10.1053/j.gastro.2015.06.043
- 59Nayak RR, Alexander M, Deshpande I, et al. Methotrexate impacts conserved pathways in diverse human gut bacteria leading to decreased host immune activation. Cell Host & Microbe. 2021;29:362-377.e11. doi:10.1016/j.chom.2020.12.008
- 60PEARL J. Causal diagrams for empirical research. Biometrika. 1995;82:669–88. doi:10.1093/biomet/82.4.669
- 61Joice R, Yasuda K, Shafquat A, et al. Determining Microbial Products and Identifying Molecular Targets in the Human Microbiome. Cell Metabolism. 2014;20:731–41. doi:10.1016/j.cmet.2014.10.003
- 62Scheline R. Metabolism of foreign compounds by gastrointestinal microorganisms. Pharmacol Rev 1973;25:451–523.https://www.ncbi.nlm.nih.gov/pubmed/4587548
- 63Klünemann M, Andrejev S, Blasche S, et al. Bioaccumulation of therapeutic drugs by human gut bacteria. Nature. 2021;597:533–8. doi:10.1038/s41586-021-03891-8
- 64Richter MF, Drown BS, Riley AP, et al. Predictive compound accumulation rules yield a broad-spectrum antibiotic. Nature. 2017;545:299–304. doi:10.1038/nature22308
- 65Haiser HJ, Seim KL, Balskus EP, et al. Mechanistic insight into digoxin inactivation byEggerthella lentaaugments our understanding of its pharmacokinetics. Gut Microbes. 2014;5:233–8. doi:10.4161/gmic.27915
- 66Maini Rekdal V, Bess EN, Bisanz JE, et al. Discovery and inhibition of an interspecies gut bacterial pathway for Levodopa metabolism. Science. 2019;364. doi:10.1126/science.aau6323
- 67Guo Y, Crnkovic CM, Won K-J, et al. Commensal Gut Bacteria Convert the Immunosuppressant Tacrolimus to Less Potent Metabolites. Drug Metab Dispos. 2018;47:194–202. doi:10.1124/dmd.118.084772
- 68Okuda H, Ogura K, Kato A, et al. A possible mechanism of eighteen patient deaths caused by interactions of sorivudine, a new antiviral drug, with oral 5-fluorouracil prodrugs. J Pharmacol Exp Ther 1998;287:791–9.https://www.ncbi.nlm.nih.gov/pubmed/9808711
- 69Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, et al. Separating host and microbiome contributions to drug pharmacokinetics and toxicity. Science. 2019;363. doi:10.1126/science.aat9931
- 70Javdan B, Lopez JG, Chankhamjon P, et al. Personalized Mapping of Drug Metabolism by the Human Gut Microbiome. Cell. 2020;181:1661-1679.e22. doi:10.1016/j.cell.2020.05.001
- 71Fuller AT. IS p-AMINOBENZENESULPHONAMIDE THE ACTIVE AGENT IN PRONTOSIL THERAPY ? The Lancet. 1937;229:194–8. doi:10.1016/s0140-6736(00)97447-6
- 72Yadav V, Gaisford S, Merchant HA, et al. Colonic bacterial metabolism of corticosteroids. International Journal of Pharmaceutics. 2013;457:268–74. doi:10.1016/j.ijpharm.2013.09.007
- 73Basit AW, Newton JM, Lacey LF. Susceptibility of the H2-receptor antagonists cimetidine, famotidine and nizatidine, to metabolism by the gastrointestinal microflora. International Journal of Pharmaceutics. 2002;237:23–33. doi:10.1016/s0378-5173(02)00018-2
- 74Wang J, Yadav V, Smart AL, et al. Stability of peptide drugs in the colon. European Journal of Pharmaceutical Sciences. 2015;78:31–6. doi:10.1016/j.ejps.2015.06.018
- 75Coombes Z, Yadav V, McCoubrey L, et al. Progestogens Are Metabolized by the Gut Microbiota: Implications for Colonic Drug Delivery. Pharmaceutics. 2020;12:760. doi:10.3390/pharmaceutics12080760
- 76Zimmermann M, Zimmermann-Kogadeeva M, Wegmann R, et al. Mapping human microbiome drug metabolism by gut bacteria and their genes. Nature. 2019;570:462–7. doi:10.1038/s41586-019-1291-3
- 77van Kessel SP, Frye AK, El-Gendy AO, et al. Gut bacterial tyrosine decarboxylases restrict levels of levodopa in the treatment of Parkinson’s disease. Nat Commun. 2019;10. doi:10.1038/s41467-019-08294-y
- 78Xu J, Liang R, Zhang W, et al. Faecalibacterium prausnitzii ‐derived microbial anti‐inflammatory molecule regulates intestinal integrity in diabetes mellitus mice via modulating tight junction protein expression. Journal of Diabetes. 2019;12:224–36. doi:10.1111/1753-0407.12986
- 79Li H, Gong Y, Xie Y, et al. Clostridium butyricum protects the epithelial barrier by maintaining tight junction protein expression and regulating microflora in a murine model of dextran sodium sulfate-induced colitis. Scandinavian Journal of Gastroenterology. 2018;53:1031–42. doi:10.1080/00365521.2016.1192678
- 80Chelakkot C, Choi Y, Kim D-K, et al. Akkermansia muciniphila-derived extracellular vesicles influence gut permeability through the regulation of tight junctions. Exp Mol Med. 2018;50:e450–e450. doi:10.1038/emm.2017.282
- 81Ghyselinck J, Verstrepen L, Moens F, et al. A 4-strain probiotic supplement influences gut microbiota composition and gut wall function in patients with ulcerative colitis. International Journal of Pharmaceutics. 2020;587:119648. doi:10.1016/j.ijpharm.2020.119648
- 82Bäsler K, Bergmann S, Heisig M, et al. The role of tight junctions in skin barrier function and dermal absorption. Journal of Controlled Release. 2016;242:105–18. doi:10.1016/j.jconrel.2016.08.007
- 83Takashima S, Tanaka F, Kawaguchi Y, et al. Proton pump inhibitors enhance intestinal permeability via dysbiosis of gut microbiota under stressed conditions in mice. Neurogastroenterology & Motility. 2020;32. doi:10.1111/nmo.13841
- 84Chen K, Zhao H, Shu L, et al. Effect of resveratrol on intestinal tight junction proteins and the gut microbiome in high-fat diet-fed insulin resistant mice. International Journal of Food Sciences and Nutrition. 2020;71:965–78. doi:10.1080/09637486.2020.1754351
- 85Tscheik C, Blasig IE, Winkler L. Trends in drug delivery through tissue barriers containing tight junctions. Tissue Barriers. 2013;1:e24565. doi:10.4161/tisb.24565
- 86González-Sarrías A, Miguel V, Merino G, et al. The Gut Microbiota Ellagic Acid-Derived Metabolite Urolithin A and Its Sulfate Conjugate Are Substrates for the Drug Efflux Transporter Breast Cancer Resistance Protein (ABCG2/BCRP). J. Agric. Food Chem. 2013;61:4352–9. doi:10.1021/jf4007505
- 87Roager HM, Hansen LBS, Bahl MI, et al. Colonic transit time is related to bacterial metabolism and mucosal turnover in the gut. Nat Microbiol. 2016;1. doi:10.1038/nmicrobiol.2016.93
- 88Ridlon JM, Kang DJ, Hylemon PB, et al. Bile acids and the gut microbiome. Current Opinion in Gastroenterology. 2014;30:332–8. doi:10.1097/mog.0000000000000057
- 89Enright EF, Griffin BT, Gahan CGM, et al. Microbiome-mediated bile acid modification: Role in intestinal drug absorption and metabolism. Pharmacological Research. 2018;133:170–86. doi:10.1016/j.phrs.2018.04.009
- 90Enright EF, Joyce SA, Gahan CGM, et al. Impact of Gut Microbiota-Mediated Bile Acid Metabolism on the Solubilization Capacity of Bile Salt Micelles and Drug Solubility. Mol. Pharmaceutics. 2017;14:1251–63. doi:10.1021/acs.molpharmaceut.6b01155
- 91Ibarra M, Trocóniz IF, Fagiolino P. Enteric reabsorption processes and their impact on drug pharmacokinetics. Sci Rep. 2021;11. doi:10.1038/s41598-021-85174-w
- 92Montagner A, Korecka A, Polizzi A, et al. Hepatic circadian clock oscillators and nuclear receptors integrate microbiome-derived signals. Sci Rep. 2016;6. doi:10.1038/srep20127
- 93Gao B, Duan Y, Lang S, et al. Functional Microbiomics Reveals Alterations of the Gut Microbiome and Host Co‐Metabolism in Patients With Alcoholic Hepatitis. Hepatol Commun. 2020;4:1168–82. doi:10.1002/hep4.1537
- 94Jones A. The health gut guide: how your diet is affecting your mental health. The Telegraph. 2020.https://www.telegraph.co.uk/beauty/body/diet-affecting-mental-health/ (accessed Feb 2022).
- 95Chadwick J. Gut bacteria in healthy old people becomes ‘increasingly unique’ as they age as their microbiome produces life-extending chemicals. The Daily Mail. 2021.https://www.dailymail.co.uk/sciencetech/article-9313731/Scientists-predict-microbiome-live-long-healthy-life.html (accessed Feb 2022).
- 96Zoe: Understand how your body responds to food. Zoe. https://joinzoe.com (accessed Feb 2022).
- 97Market data: The Probiotic market overview. International Probiotics Association. 2021.https://www.ipaeurope.org/legal-framework/market-data/#:~:text=The%20global%20probiotics%20market%2C%20for,for%20the%20period%202019%2D2024 (accessed Feb 2022).
- 98Veiga P, Suez J, Derrien M, et al. Moving from probiotics to precision probiotics. Nat Microbiol. 2020;5:878–80. doi:10.1038/s41564-020-0721-1
- 99Suez J, Zmora N, Zilberman-Schapira G, et al. Post-Antibiotic Gut Mucosal Microbiome Reconstitution Is Impaired by Probiotics and Improved by Autologous FMT. Cell. 2018;174:1406-1423.e16. doi:10.1016/j.cell.2018.08.047
- 100Sniffen JC, McFarland LV, Evans CT, et al. Choosing an appropriate probiotic product for your patient: An evidence-based practical guide. PLoS ONE. 2018;13:e0209205. doi:10.1371/journal.pone.0209205
- 101Fredua-Agyeman M, Gaisford S. Comparative survival of commercial probiotic formulations: tests in biorelevant gastric fluids and real-time measurements using microcalorimetry. Beneficial Microbes. 2015;6:141–51. doi:10.3920/bm2014.0051
- 102Dodoo CC, Wang J, Basit AW, et al. Targeted delivery of probiotics to enhance gastrointestinal stability and intestinal colonisation. International Journal of Pharmaceutics. 2017;530:224–9. doi:10.1016/j.ijpharm.2017.07.068
- 103McFarland LV. Meta-Analysis of Probiotics for the Prevention of Antibiotic Associated Diarrhea and the Treatment of Clostridium difficile Disease. Am J Gastroenterol. 2006;101:812–22. doi:10.1111/j.1572-0241.2006.00465.x
- 104Buffie CG, Bucci V, Stein RR, et al. Precision microbiome reconstitution restores bile acid mediated resistance to Clostridium difficile. Nature. 2014;517:205–8. doi:10.1038/nature13828
- 105McFarland LV. Efficacy of Single-Strain Probiotics Versus Multi-Strain Mixtures: Systematic Review of Strain and Disease Specificity. Dig Dis Sci. 2020;66:694–704. doi:10.1007/s10620-020-06244-z
- 106Antimicrobial stewardship: systems and processes for effective antimicrobial medicine use NICE guideline [NG15]. National Institute for Health and Care Excellence. 2015.https://www.nice.org.uk/guidance/ng15 (accessed Feb 2022).
- 107Vilcu A-M, Sabatte L, Blanchon T, et al. Association Between Acute Gastroenteritis and Continuous Use of Proton Pump Inhibitors During Winter Periods of Highest Circulation of Enteric Viruses. JAMA Netw Open. 2019;2:e1916205. doi:10.1001/jamanetworkopen.2019.16205
- 108Mallory EK, Acharya A, Rensi SE, et al. Chemical reaction vector embeddings: towards predicting drug metabolism in the human gut microbiome. Biocomputing 2018. 2017. doi:10.1142/9789813235533_0006
- 109McCoubrey LE, Elbadawi M, Orlu M, et al. Machine Learning Uncovers Adverse Drug Effects on Intestinal Bacteria. Pharmaceutics. 2021;13:1026. doi:10.3390/pharmaceutics13071026
- 110McCoubrey LE, Thomaidou S, Elbadawi M, et al. Machine Learning Predicts Drug Metabolism and Bioaccumulation by Intestinal Microbiota. Pharmaceutics. 2021;13:2001. doi:10.3390/pharmaceutics13122001
- 111Wilkinson JE, Franzosa EA, et al. A framework for microbiome science in public health. Nat Med. 2021;27:766–74. doi:10.1038/s41591-021-01258-0
- 112Van Norman GA. Drugs, Devices, and the FDA: Part 1. JACC: Basic to Translational Science. 2016;1:170–9. doi:10.1016/j.jacbts.2016.03.002
- 113ICH guideline M3(R2) on non-clinical safety studies for the conduct of human clinical trials and marketing authorisation for pharmaceuticals. European Medicines Agency. 2009.https://www.ema.europa.eu/en/ich-m3-r2-non-clinical-safety-studies-conduct-human-clinical-trials-pharmaceuticals (accessed Feb 2022).
- 114McCoubrey LE, Elbadawi M, Orlu M, et al. Harnessing machine learning for development of microbiome therapeutics. Gut Microbes. 2021;13. doi:10.1080/19490976.2021.1872323
- 115Prattichizzo F, Giuliani A, Mensà E, et al. Pleiotropic effects of metformin: Shaping the microbiome to manage type 2 diabetes and postpone ageing. Ageing Research Reviews. 2018;48:87–98. doi:10.1016/j.arr.2018.10.003
- 116Tannergren C, Borde A, Boreström C, et al. Evaluation of an in vitro faecal degradation method for early assessment of the impact of colonic degradation on colonic absorption in humans. European Journal of Pharmaceutical Sciences. 2014;57:200–6. doi:10.1016/j.ejps.2013.10.001
- 117Awad A, Madla CM, McCoubrey LE, et al. Clinical translation of advanced colonic drug delivery technologies. Advanced Drug Delivery Reviews. 2022;181:114076. doi:10.1016/j.addr.2021.114076
- 118Varum F, Freire AC, Bravo R, et al. OPTICORETM, an innovative and accurate colonic targeting technology. International Journal of Pharmaceutics. 2020;583:119372. doi:10.1016/j.ijpharm.2020.119372
- 119Varum F, Freire AC, Fadda HM, et al. A dual pH and microbiota-triggered coating (PhloralTM) for fail-safe colonic drug release. International Journal of Pharmaceutics. 2020;583:119379. doi:10.1016/j.ijpharm.2020.119379
- 120IBEKWE VC, KHELA MK, EVANS DF, et al. A new concept in colonic drug targeting: a combined pH-responsive and bacterially-triggered drug delivery technology. Alimentary Pharmacology & Therapeutics. 2008;28:911–6. doi:10.1111/j.1365-2036.2008.03810.x